You are browsing environment: FUNGIDB
CAZyme Information: KAG1706915.1
You are here: Home > Sequence: KAG1706915.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1706915.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.122:14 | - |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 136 | 336 | 1.5e-36 | 0.9791666666666666 |
| GT10 | 650 | 803 | 1.6e-35 | 0.41210374639769454 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 2.03e-27 | 136 | 338 | 3 | 196 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 395683 | Glyco_transf_10 | 1.42e-13 | 673 | 797 | 17 | 130 | Glycosyltransferase family 10 (fucosyltransferase) C-term. This is the C-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The C-terminal domain is the likely binding-region for ADP (manuscript in publication). |
| 215596 | PLN03133 | 3.32e-09 | 121 | 327 | 387 | 579 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.24e-19 | 595 | 804 | 14 | 211 | |
| 4.13e-19 | 583 | 821 | 370 | 597 | |
| 4.45e-19 | 653 | 804 | 120 | 265 | |
| 4.45e-19 | 653 | 804 | 120 | 265 | |
| 4.45e-19 | 653 | 804 | 120 | 265 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.30e-15 | 120 | 358 | 110 | 339 | Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMN_B Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMN_C Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMN_D Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMO_A Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMO_B Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens] |
|
| 5.59e-15 | 120 | 358 | 116 | 345 | Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHK_B Chain B, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHK_C Chain C, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHK_D Chain D, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHL_A Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHL_B Chain B, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHM_A Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHM_B Chain B, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHN_A Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHO_A Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHO_B Chain B, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 6.30e-15 | 120 | 358 | 130 | 359 | Chain A, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHI_B Chain B, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHI_C Chain C, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens],7JHI_D Chain D, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 1.27e-14 | 120 | 358 | 110 | 339 | Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens],6WMM_B Human poly-N-acetyl-lactosamine synthase structure demonstrates a modular assembly of catalytic subsites for GT-A glycosyltransferases [Homo sapiens] |
|
| 1.02e-12 | 658 | 809 | 179 | 316 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_C Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZX_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZY_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.63e-19 | 118 | 325 | 85 | 287 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Sus scrofa OX=9823 GN=B3GNT5 PE=2 SV=1 |
|
| 8.88e-19 | 595 | 804 | 369 | 566 | Putative fucosyltransferase R654 OS=Acanthamoeba polyphaga mimivirus OX=212035 GN=MIMI_R654 PE=3 SV=1 |
|
| 1.20e-17 | 118 | 325 | 87 | 288 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Homo sapiens OX=9606 GN=B3GNT5 PE=1 SV=1 |
|
| 1.22e-16 | 118 | 325 | 85 | 286 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Mus musculus OX=10090 GN=B3gnt5 PE=2 SV=1 |
|
| 1.02e-15 | 120 | 339 | 56 | 260 | Beta-1,3-galactosyltransferase 5 OS=Mus musculus OX=10090 GN=B3galt5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
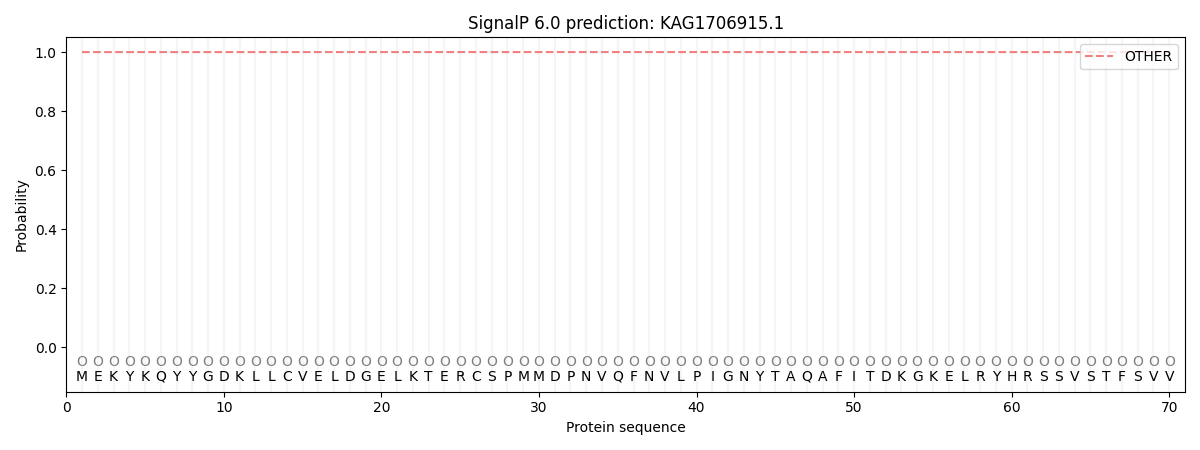
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000042 | 0.000001 |
