You are browsing environment: FUNGIDB
CAZyme Information: KAG1703273.1
You are here: Home > Sequence: KAG1703273.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1703273.1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 109 | 316 | 6.4e-63 | 0.9120370370370371 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185053 | PRK15098 | 3.34e-107 | 42 | 741 | 51 | 754 | beta-glucosidase BglX. |
| 224389 | BglX | 3.76e-68 | 112 | 440 | 58 | 373 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| 178629 | PLN03080 | 2.11e-48 | 129 | 708 | 108 | 742 | Probable beta-xylosidase; Provisional |
| 395747 | Glyco_hydro_3 | 4.46e-47 | 42 | 335 | 6 | 287 | Glycosyl hydrolase family 3 N terminal domain. |
| 396478 | Glyco_hydro_3_C | 4.77e-43 | 403 | 629 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 764 | 1 | 767 | |
| 0.0 | 5 | 760 | 50 | 820 | |
| 0.0 | 5 | 760 | 5 | 764 | |
| 7.63e-291 | 45 | 759 | 1 | 724 | |
| 2.46e-288 | 28 | 759 | 67 | 808 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.58e-128 | 19 | 745 | 7 | 746 | Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus],5JP0_B Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus] |
|
| 1.39e-86 | 32 | 605 | 19 | 587 | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X38_A crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X39_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare] |
|
| 1.50e-86 | 32 | 605 | 19 | 587 | Beta-d-glucan Exohydrolase From Barley [Hordeum vulgare],1IEQ_A Crystal Structure Of Barley Beta-D-Glucan Glucohydrolase Isoenzyme Exo1 [Hordeum vulgare],1IEV_A Crystal Structure Of Barley Beta-d-glucan Glucohydrolase Isoenzyme Exo1 In Complex With Cyclohexitol [Hordeum vulgare],1IEW_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside [Hordeum vulgare],1IEX_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose [Hordeum vulgare],1J8V_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside [Hordeum vulgare],3WLH_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLJ_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 3-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLK_X Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with 4-deoxy-glucose [Hordeum vulgare subsp. vulgare],3WLL_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with PEG400 [Hordeum vulgare subsp. vulgare],3WLM_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with octyl-O-glucoside [Hordeum vulgare subsp. vulgare],3WLN_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme EXO1 in complex with octyl-S-glucoside [Hordeum vulgare subsp. vulgare],3WLP_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare] |
|
| 1.66e-86 | 32 | 605 | 23 | 591 | Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],3WLO_A Crystal Structure Analysis of Plant Exohydrolase [Hordeum vulgare subsp. vulgare],6JG1_A Chain A, Barley exohydrolase I [Hordeum vulgare subsp. vulgare],6JG2_A Chain A, Barley exohydrolase I [Hordeum vulgare subsp. vulgare],6MD6_A CRYSTAL STRUCTURE ANALYSIS OF PLANT EXOHYDROLASE IN COMPLEX WITH METHYL 2-THIO-BETA-SOPHOROSIDE [Hordeum vulgare subsp. vulgare] |
|
| 3.27e-85 | 32 | 605 | 23 | 591 | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 2-thio-beta-sophoroside. [Hordeum vulgare subsp. vulgare],6JGR_A Crystal structure of barley exohydrolaseI W434Y in complex with 4'-nitrophenyl thiolaminaribioside [Hordeum vulgare subsp. vulgare],6JGS_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. [Hordeum vulgare subsp. vulgare],6JGT_A Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. [Hordeum vulgare subsp. vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.11e-144 | 32 | 737 | 83 | 811 | Lysosomal beta glucosidase OS=Dictyostelium discoideum OX=44689 GN=gluA PE=1 SV=2 |
|
| 2.67e-127 | 19 | 745 | 29 | 768 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
|
| 8.38e-88 | 99 | 750 | 50 | 719 | Beta-xylosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22130 PE=1 SV=1 |
|
| 3.93e-85 | 115 | 720 | 105 | 737 | Periplasmic beta-glucosidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=bglX PE=3 SV=2 |
|
| 1.46e-84 | 26 | 720 | 35 | 737 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
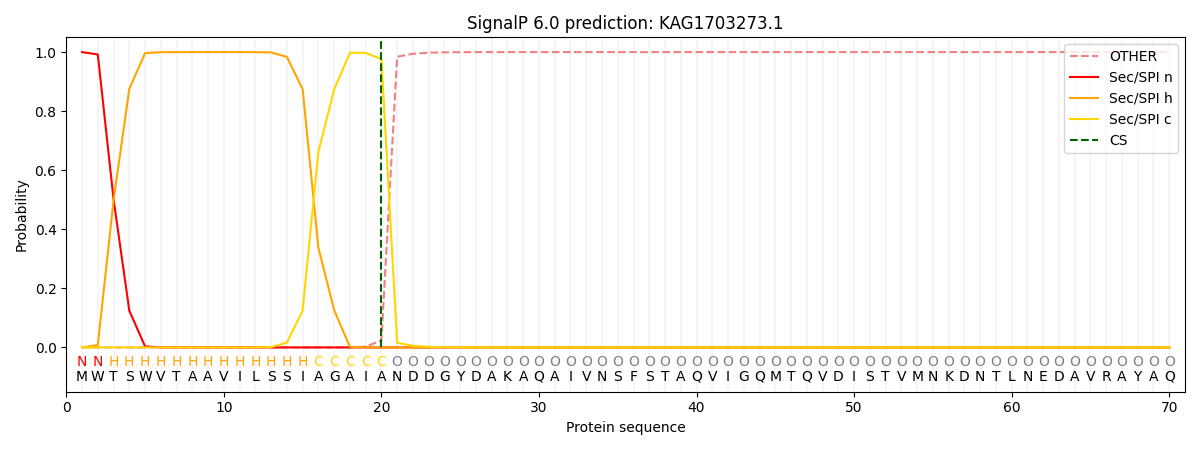
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000223 | 0.999749 | CS pos: 20-21. Pr: 0.9765 |
