You are browsing environment: FUNGIDB
CAZyme Information: KAG1699936.1
You are here: Home > Sequence: KAG1699936.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1699936.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 262 | 5.7e-57 | 0.9836734693877551 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411418 | ser_rich_anae_1 | 6.48e-04 | 276 | 393 | 316 | 432 | serine-rich protein. This serine-rich protein belongs to a family with large size (over 1000 amino acids), which a highly serine-rich central region that averages over 300 aa in length. Species encoding members of this family of proteins tend to be anaerobic bacteria, including Gram-positive bacteria of the human gut microbiome and Chloroflexi from marine sediments. |
| 411418 | ser_rich_anae_1 | 0.001 | 276 | 399 | 254 | 384 | serine-rich protein. This serine-rich protein belongs to a family with large size (over 1000 amino acids), which a highly serine-rich central region that averages over 300 aa in length. Species encoding members of this family of proteins tend to be anaerobic bacteria, including Gram-positive bacteria of the human gut microbiome and Chloroflexi from marine sediments. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.37e-107 | 7 | 197 | 7 | 197 | |
| 2.67e-104 | 1 | 197 | 1 | 197 | |
| 3.78e-104 | 7 | 197 | 7 | 197 | |
| 1.06e-77 | 1 | 197 | 1 | 174 | |
| 1.47e-71 | 13 | 197 | 14 | 202 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-13 | 101 | 195 | 69 | 174 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
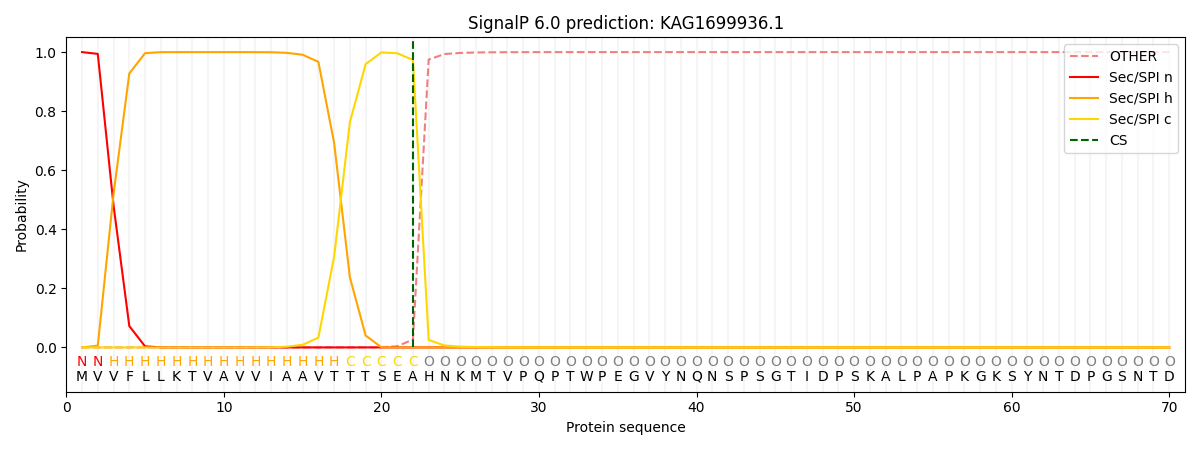
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000212 | 0.999789 | CS pos: 22-23. Pr: 0.9735 |
