You are browsing environment: FUNGIDB
CAZyme Information: KAG1692718.1
You are here: Home > Sequence: KAG1692718.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
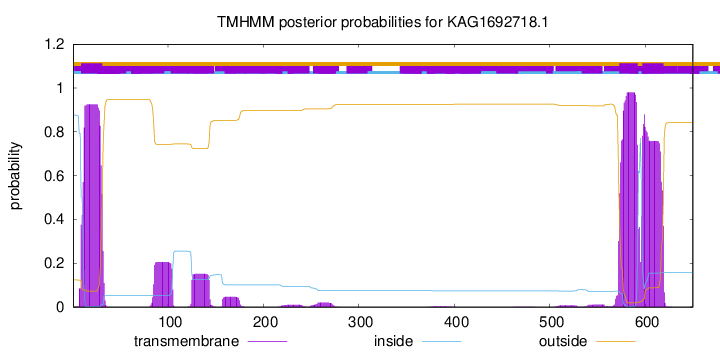
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1692718.1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 124 | 551 | 1.4e-26 | 0.8952879581151832 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 1.02e-46 | 85 | 545 | 2 | 398 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 1.15e-27 | 85 | 554 | 3 | 400 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 278624 | UDPGT | 4.79e-17 | 167 | 590 | 93 | 477 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 177813 | PLN02152 | 4.59e-10 | 85 | 505 | 5 | 397 | indole-3-acetate beta-glucosyltransferase |
| 215127 | PLN02210 | 2.90e-08 | 82 | 543 | 7 | 442 | UDP-glucosyl transferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 626 | 1 | 632 | |
| 1.01e-161 | 84 | 606 | 64 | 548 | |
| 4.56e-65 | 83 | 553 | 4 | 424 | |
| 8.20e-64 | 83 | 553 | 2 | 421 | |
| 1.63e-52 | 83 | 550 | 2 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.28e-14 | 85 | 506 | 10 | 424 | Crystal structure of Medicago truncatula UGT85H2- Insights into the structural basis of a multifunctional (Iso) flavonoid glycosyltransferase [Medicago truncatula] |
|
| 8.28e-09 | 83 | 554 | 5 | 443 | Crystal structure of a flavonoid C-glucosyltrasferase from Fagopyrum esculentum (FeCGTa) complexed with BrUTP [Fagopyrum esculentum] |
|
| 8.56e-09 | 83 | 554 | 19 | 457 | Crystal Structure of Fagopyrum esculentum M UGT708C1 [Fagopyrum esculentum],6LLG_B Crystal Structure of Fagopyrum esculentum M UGT708C1 [Fagopyrum esculentum],6LLW_A Crystal Structure of Fagopyrum esculentum M UGT708C1 complexed with UDP [Fagopyrum esculentum],6LLW_B Crystal Structure of Fagopyrum esculentum M UGT708C1 complexed with UDP [Fagopyrum esculentum],6LLZ_A Crystal Structure of Fagopyrum esculentum M UGT708C1 complexed with UDP-glucose [Fagopyrum esculentum],6LLZ_B Crystal Structure of Fagopyrum esculentum M UGT708C1 complexed with UDP-glucose [Fagopyrum esculentum] |
|
| 1.33e-08 | 86 | 549 | 23 | 403 | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form [Micromonospora echinospora],3OTH_A Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form [Micromonospora echinospora],3OTH_B Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form [Micromonospora echinospora] |
|
| 3.30e-08 | 453 | 554 | 332 | 433 | Structural Characterization of UDP-glycosyltransferase from Tetranychus Urticae [Tetranychus urticae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.68e-18 | 157 | 616 | 91 | 512 | UDP-glycosyltransferase UGT5 OS=Dactylopius coccus OX=765876 GN=UGT5 PE=1 SV=1 |
|
| 2.83e-17 | 197 | 606 | 139 | 511 | UDP-glucuronosyltransferase 3A2 OS=Mus musculus OX=10090 GN=Ugt3a2 PE=1 SV=2 |
|
| 4.98e-17 | 197 | 577 | 139 | 488 | UDP-glucuronosyltransferase 3A1 OS=Mus musculus OX=10090 GN=Ugt3a1 PE=1 SV=1 |
|
| 2.89e-16 | 86 | 577 | 24 | 477 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase OS=Rattus norvegicus OX=10116 GN=Ugt8 PE=1 SV=1 |
|
| 2.89e-16 | 86 | 604 | 24 | 493 | 2-hydroxyacylsphingosine 1-beta-galactosyltransferase OS=Homo sapiens OX=9606 GN=UGT8 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.438988 | 0.561001 | CS pos: 29-30. Pr: 0.5282 |