You are browsing environment: FUNGIDB
CAZyme Information: KAG1688503.1
You are here: Home > Sequence: KAG1688503.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1688503.1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.109:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT105 | 82 | 247 | 2.9e-32 | 0.9389312977099237 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400627 | DUF1736 | 1.14e-18 | 293 | 356 | 11 | 74 | Domain of unknown function (DUF1736). This domain of unknown function is found in various hypothetical metazoan proteins. |
| 276809 | TPR | 8.94e-12 | 634 | 739 | 4 | 96 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 1.69e-09 | 616 | 706 | 20 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 223533 | TPR | 1.92e-08 | 512 | 739 | 71 | 263 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
| 276809 | TPR | 2.43e-05 | 680 | 739 | 3 | 62 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.59e-65 | 4 | 513 | 15 | 492 | |
| 4.61e-65 | 4 | 526 | 14 | 506 | |
| 2.03e-64 | 4 | 513 | 15 | 492 | |
| 5.26e-64 | 4 | 526 | 14 | 506 | |
| 5.26e-64 | 4 | 526 | 14 | 506 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.82e-66 | 4 | 513 | 15 | 492 | Protein O-mannosyl-transferase TMTC4 OS=Homo sapiens OX=9606 GN=TMTC4 PE=1 SV=2 |
|
| 9.34e-65 | 4 | 526 | 14 | 506 | Protein O-mannosyl-transferase TMTC4 OS=Mus musculus OX=10090 GN=Tmtc4 PE=2 SV=1 |
|
| 9.50e-60 | 4 | 580 | 8 | 525 | Protein O-mannosyl-transferase TMTC3 OS=Mus musculus OX=10090 GN=Tmtc3 PE=1 SV=2 |
|
| 7.74e-59 | 14 | 580 | 11 | 520 | Protein O-mannosyl-transferase TMTC3 OS=Homo sapiens OX=9606 GN=TMTC3 PE=1 SV=2 |
|
| 5.97e-57 | 6 | 561 | 29 | 572 | Protein O-mannosyl-transferase Tmtc3 OS=Drosophila melanogaster OX=7227 GN=Tmtc3 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000609 | 0.999361 | CS pos: 30-31. Pr: 0.8705 |
TMHMM Annotations download full data without filtering help
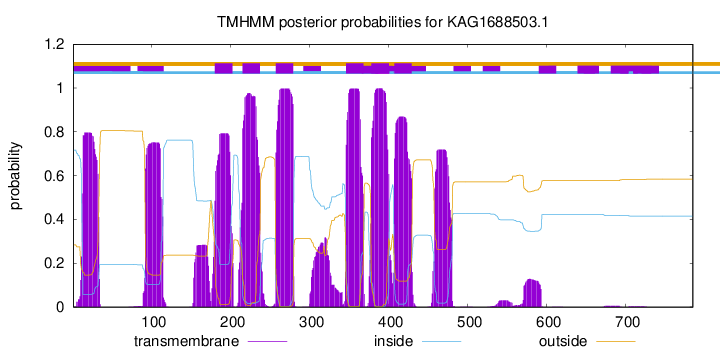
| Start | End |
|---|---|
| 180 | 202 |
| 215 | 237 |
| 257 | 279 |
| 346 | 368 |
| 378 | 400 |
| 407 | 429 |
