You are browsing environment: FUNGIDB
CAZyme Information: KAG1687351.1
You are here: Home > Sequence: KAG1687351.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1687351.1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 10 | 256 | 2.8e-66 | 0.9346938775510204 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.79e-142 | 1 | 213 | 1 | 214 | |
| 5.79e-142 | 1 | 213 | 1 | 214 | |
| 5.49e-141 | 1 | 213 | 1 | 214 | |
| 5.49e-141 | 1 | 213 | 1 | 214 | |
| 1.06e-129 | 1 | 200 | 1 | 201 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.35e-91 | 23 | 197 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
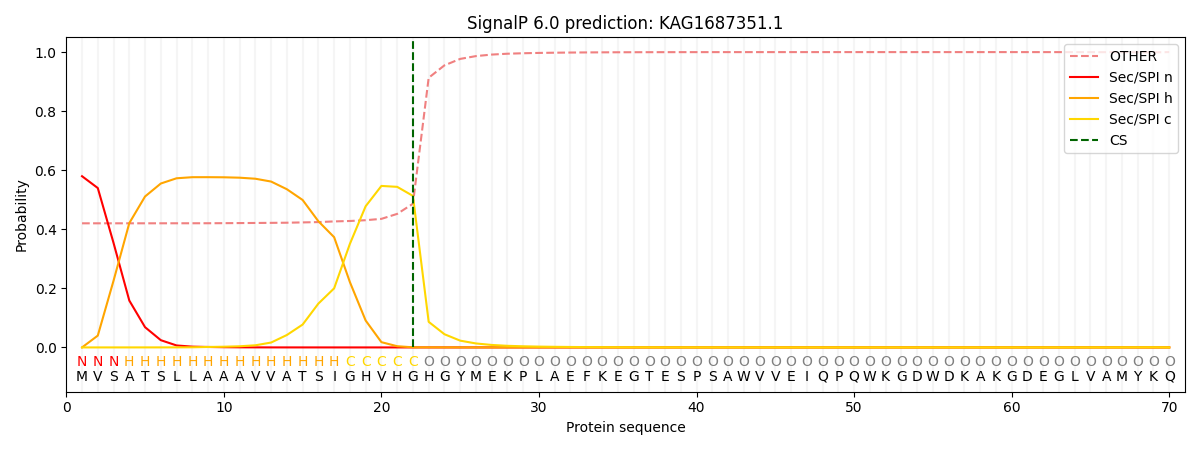
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.433467 | 0.566535 | CS pos: 22-23. Pr: 0.5131 |
