You are browsing environment: FUNGIDB
CAZyme Information: KAF7629670.1
You are here: Home > Sequence: KAF7629670.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus flavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus flavus | |||||||||||
| CAZyme ID | KAF7629670.1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase [Source:UniProtKB/TrEMBL;Acc:B8NYB7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 70 | 243 | 2e-49 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 2.99e-62 | 69 | 243 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.83e-184 | 1 | 248 | 1 | 248 | |
| 7.83e-184 | 1 | 248 | 1 | 248 | |
| 7.83e-184 | 1 | 248 | 1 | 248 | |
| 7.83e-184 | 1 | 248 | 1 | 248 | |
| 7.83e-184 | 1 | 248 | 1 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.45e-46 | 42 | 246 | 6 | 206 | Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus],3WP3_B Xylanase 11C from Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) [Talaromyces funiculosus] |
|
| 9.92e-46 | 61 | 246 | 5 | 189 | Crystal structure of family 11 xylanase in complex with inhibitor (XIP-I) [Talaromyces funiculosus] |
|
| 1.94e-43 | 70 | 246 | 15 | 192 | High resolution structure of GH11 xylanase from Nectria haematococca [Fusarium vanettenii 77-13-4] |
|
| 3.55e-43 | 70 | 246 | 13 | 189 | Chain A, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_B Chain B, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_C Chain C, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_D Chain D, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_E Chain E, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_F Chain F, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_G Chain G, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_H Chain H, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_I Chain I, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_J Chain J, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_K Chain K, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612],5HXV_L Chain L, Endo-1,4-beta-xylanase [Talaromyces cellulolyticus CF-2612] |
|
| 4.47e-42 | 68 | 248 | 14 | 194 | Crystal Structure of a Xylanase at 1.56 Angstroem resolution [Fusarium oxysporum],5JRN_A Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution [Fusarium oxysporum f. sp. vasinfectum 25433] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.32e-46 | 49 | 247 | 22 | 225 | Endo-1,4-beta-xylanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xlnB PE=3 SV=2 |
|
| 6.89e-46 | 1 | 246 | 1 | 227 | Endo-1,4-beta-xylanase xynf11a OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnA PE=1 SV=1 |
|
| 6.89e-46 | 1 | 246 | 1 | 227 | Probable endo-1,4-beta-xylanase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=xlnA PE=3 SV=1 |
|
| 1.09e-45 | 1 | 246 | 1 | 231 | Endo-1,4-beta-xylanase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xlnA PE=1 SV=1 |
|
| 1.09e-45 | 1 | 246 | 1 | 231 | Probable endo-1,4-beta-xylanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=xlnA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
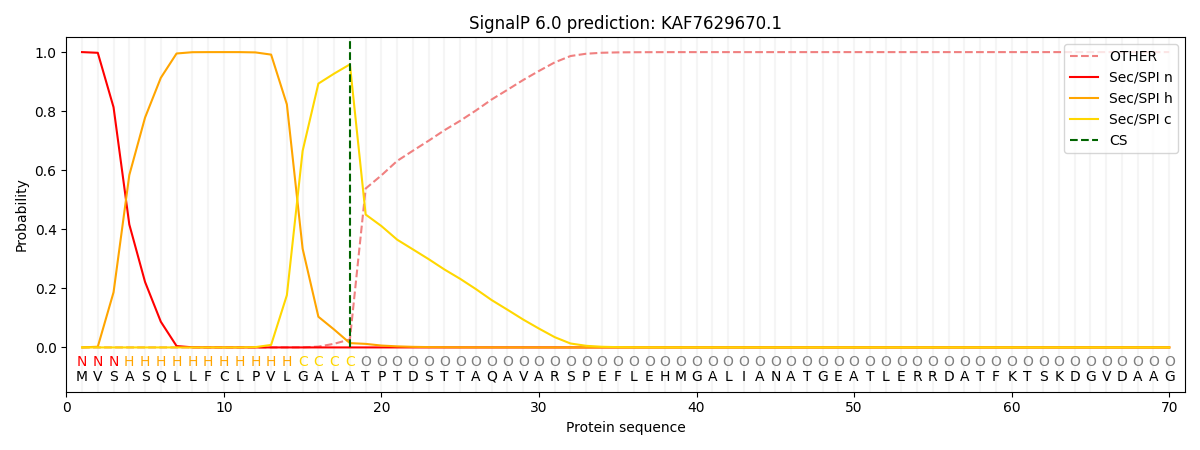
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000307 | 0.999674 | CS pos: 18-19. Pr: 0.9584 |
