You are browsing environment: FUNGIDB
CAZyme Information: KAF6528523.1
You are here: Home > Sequence: KAF6528523.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium oxysporum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium oxysporum | |||||||||||
| CAZyme ID | KAF6528523.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.4:59 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH45 | 22 | 220 | 5.8e-87 | 0.9949494949494949 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 280233 | Glyco_hydro_45 | 3.14e-112 | 21 | 220 | 1 | 214 | Glycosyl hydrolase family 45. |
| 183854 | PRK13042 | 1.35e-07 | 248 | 321 | 24 | 94 | superantigen-like protein SSL4; Reviewed. |
| 197593 | fCBD | 8.55e-07 | 346 | 382 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 236940 | PRK11633 | 4.09e-06 | 221 | 318 | 50 | 146 | cell division protein DedD; Provisional |
| 411470 | opacity_OapA | 4.99e-06 | 228 | 370 | 193 | 342 | opacity-associated protein OapA. This family consists of full-length homologs to OapA, opacity-associated protein A as described in Haemophilus influenzae. OapA shares a C-terminal homology domain, called the OapA domain, with the Escherichia coli protein YtfB, which is now known to bind peptidoglycan through its OapA domain and to act as a cell division protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.39e-240 | 1 | 384 | 1 | 381 | |
| 1.39e-240 | 1 | 384 | 1 | 381 | |
| 1.39e-240 | 1 | 384 | 1 | 381 | |
| 1.39e-240 | 1 | 384 | 1 | 381 | |
| 8.63e-236 | 1 | 384 | 1 | 376 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.95e-123 | 1 | 230 | 1 | 232 | Apo Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A],6MVJ_A Cellobiose complex Cel45A from Neurospora crassa OR74A [Neurospora crassa OR74A] |
|
| 6.10e-115 | 18 | 230 | 1 | 211 | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126],5GM9_A Crystal structure of a glycoside hydrolase in complex with cellobiose [Thermothielavioides terrestris NRRL 8126] |
|
| 7.02e-115 | 18 | 230 | 4 | 214 | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 [Thermothielavioides terrestris NRRL 8126] |
|
| 5.18e-109 | 21 | 231 | 2 | 210 | Structure of Melanocarpus albomyces endoglucanase in complex with cellobiose [Melanocarpus albomyces],1OA9_A Structure of Melanocarpus albomyces endoglucanase [Melanocarpus albomyces] |
|
| 1.33e-107 | 21 | 227 | 2 | 206 | Structure of 20K-endoglucanase from Melanocarpus albomyces at 1.8 A [Melanocarpus albomyces] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.18e-236 | 1 | 384 | 1 | 376 | Putative endoglucanase type K OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 1.58e-104 | 22 | 231 | 3 | 211 | Endoglucanase-5 OS=Humicola insolens OX=34413 PE=1 SV=1 |
|
| 5.15e-80 | 1 | 221 | 1 | 221 | Endoglucanase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
|
| 5.08e-58 | 19 | 219 | 37 | 240 | Endoglucanase OS=Phaedon cochleariae OX=80249 PE=2 SV=1 |
|
| 4.91e-42 | 6 | 232 | 9 | 247 | Endoglucanase 1 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=EGL1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
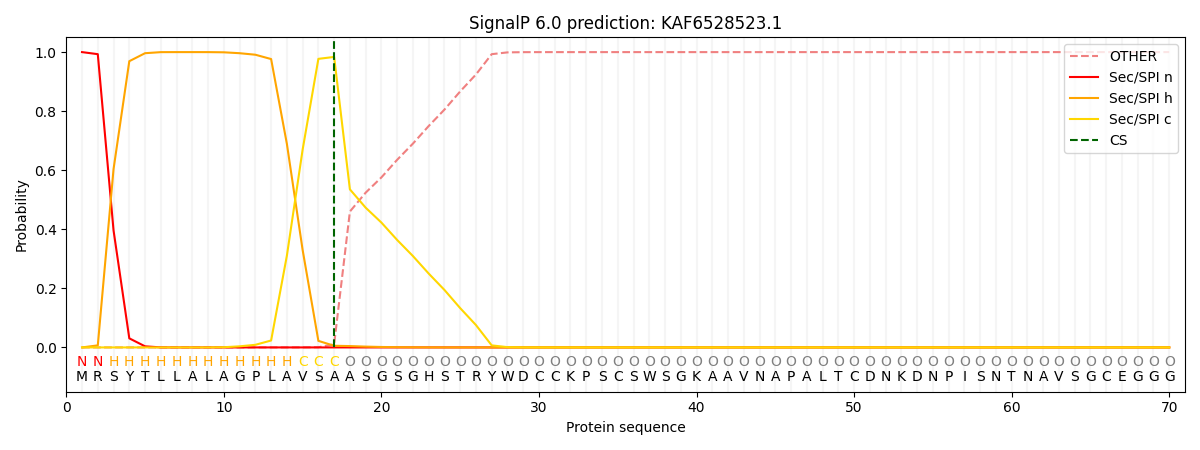
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000233 | 0.999740 | CS pos: 17-18. Pr: 0.9836 |
