You are browsing environment: FUNGIDB
CAZyme Information: KAF6516903.1
You are here: Home > Sequence: KAF6516903.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium oxysporum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium oxysporum | |||||||||||
| CAZyme ID | KAF6516903.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 406865 | AMPK1_CBM | 2.77e-15 | 5 | 53 | 4 | 50 | Glycogen recognition site of AMP-activated protein kinase. AMPK1_CBM is a family found in close association with AMPKBI pfam04739. The surface of AMPK1_CBM reveals a carbohydrate-binding pocket. |
| 199889 | E_set_AMPKbeta_like_N | 1.62e-13 | 3 | 53 | 1 | 51 | N-terminal Early set domain, a glycogen binding domain, associated with the catalytic domain of AMP-activated protein kinase beta subunit. E or "early" set domains are associated with the catalytic domain of AMP-activated protein kinase beta subunit glycogen binding domain at the N-terminal end. AMPK is a metabolic stress sensing protein that senses AMP/ATP and has recently been found to act as a glycogen sensor as well. The protein functions as an alpha-beta-gamma heterotrimer. This N-terminal domain is the glycogen binding domain of the beta subunit. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and isoamylase. |
| 173412 | PTZ00121 | 9.01e-08 | 296 | 643 | 1346 | 1690 | MAEBL; Provisional |
| 236766 | rne | 3.77e-07 | 475 | 679 | 846 | 1027 | ribonuclease E; Reviewed |
| 173412 | PTZ00121 | 4.19e-06 | 392 | 693 | 1280 | 1571 | MAEBL; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 711 | 1 | 692 | |
| 0.0 | 1 | 711 | 1 | 695 | |
| 0.0 | 1 | 711 | 1 | 695 | |
| 0.0 | 1 | 711 | 1 | 695 | |
| 0.0 | 1 | 711 | 1 | 695 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
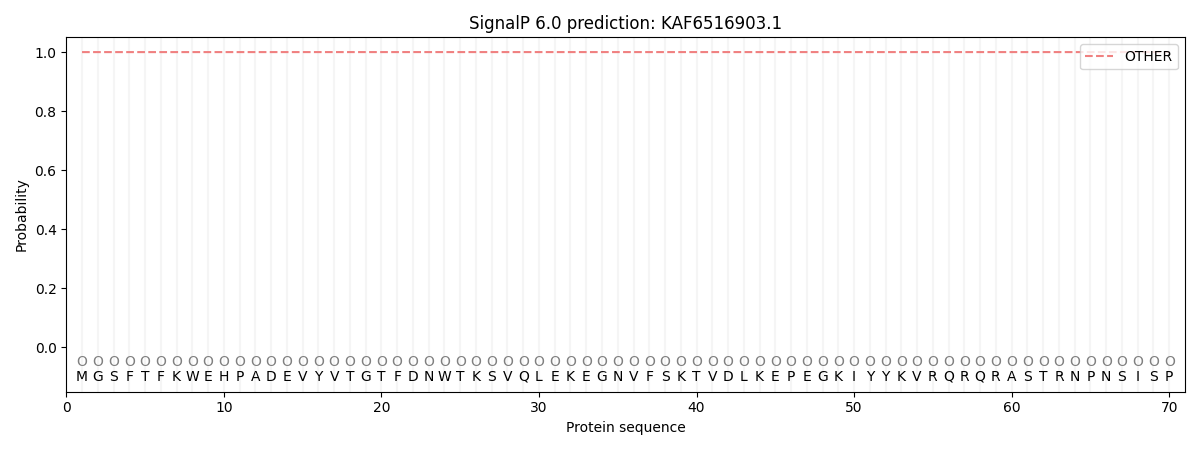
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000000 |
