You are browsing environment: FUNGIDB
CAZyme Information: KAF5684695.1
You are here: Home > Sequence: KAF5684695.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
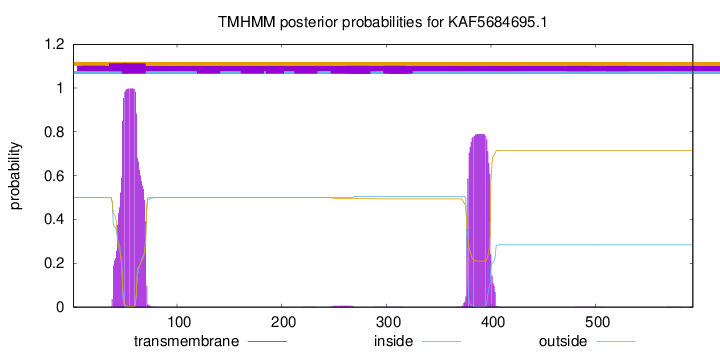
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5684695.1 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | exostosin-like 3 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT64 | 110 | 351 | 3.2e-51 | 0.9637096774193549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396528 | PTH2 | 2.74e-61 | 470 | 593 | 1 | 115 | Peptidyl-tRNA hydrolase PTH2. Peptidyl-tRNA hydrolases are enzymes that release tRNAs from peptidyl-tRNA during translation. |
| 239108 | PTH2 | 8.28e-61 | 470 | 593 | 1 | 115 | Peptidyl-tRNA hydrolase, type 2 (PTH2). Peptidyl-tRNA hydrolase (PTH) activity releases tRNA from the premature translation termination product peptidyl-tRNA, therefore allowing the tRNA and peptide to be reused in protein synthesis. PTH2 is present in archaea and eukaryotes. |
| 239091 | PTH2_family | 8.92e-55 | 470 | 593 | 1 | 115 | Peptidyl-tRNA hydrolase, type 2 (PTH2)_like . Peptidyl-tRNA hydrolase activity releases tRNA from the premature translation termination product peptidyl-tRNA. Two structurally different enzymes have been reported to encode such activity, Pth present in bacteria and eukaryotes and Pth2 present in archaea and eukaryotes. |
| 401264 | Glyco_transf_64 | 6.05e-54 | 110 | 352 | 1 | 237 | Glycosyl transferase family 64 domain. Members of this family catalyze the transfer reaction of N-acetylglucosamine and N-acetylgalactosamine from the respective UDP-sugars to the non-reducing end of [glucuronic acid]beta 1-3[galactose]beta 1-O-naphthalenemethanol, an acceptor substrate analog of the natural common linker of various glycosylaminoglycans. They are also required for the biosynthesis of heparan-sulphate. |
| 224901 | Pth2 | 1.81e-46 | 467 | 593 | 3 | 122 | Peptidyl-tRNA hydrolase [Translation, ribosomal structure and biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 593 | 1 | 527 | |
| 0.0 | 1 | 593 | 13 | 618 | |
| 1.86e-261 | 1 | 356 | 1 | 356 | |
| 1.86e-261 | 1 | 356 | 1 | 356 | |
| 1.86e-261 | 1 | 356 | 1 | 356 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.73e-30 | 469 | 592 | 2 | 116 | Crystal structure of bit1 [Homo sapiens],1Q7S_B Crystal structure of bit1 [Homo sapiens] |
|
| 1.34e-27 | 104 | 348 | 628 | 864 | Chain A, Exostosin-like 3 [Homo sapiens],7AU2_B Chain B, Exostosin-like 3 [Homo sapiens],7AUA_A Chain A, Exostosin-like 3 [Homo sapiens],7AUA_B Chain B, Exostosin-like 3 [Homo sapiens] |
|
| 4.49e-24 | 108 | 347 | 28 | 272 | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) [Mus musculus],1OMX_B Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) [Mus musculus],1OMZ_A crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc [Mus musculus],1OMZ_B crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc [Mus musculus],1ON6_A Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc [Mus musculus],1ON6_B Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc [Mus musculus],1ON8_A Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog [Mus musculus],1ON8_B Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog [Mus musculus] |
|
| 1.83e-23 | 471 | 593 | 3 | 120 | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase [Saccharolobus solfataricus P2],1XTY_B Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase [Saccharolobus solfataricus P2],1XTY_C Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase [Saccharolobus solfataricus P2],1XTY_D Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase [Saccharolobus solfataricus P2] |
|
| 1.32e-21 | 471 | 593 | 4 | 117 | Chain A, Hypothetical protein Ta0108 [Thermoplasma acidophilum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.21e-32 | 469 | 592 | 90 | 204 | Probable peptidyl-tRNA hydrolase 2 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC19A8.14 PE=1 SV=1 |
|
| 2.22e-31 | 463 | 592 | 81 | 207 | Peptidyl-tRNA hydrolase 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PTH2 PE=1 SV=2 |
|
| 2.73e-31 | 462 | 592 | 60 | 180 | Peptidyl-tRNA hydrolase 2, mitochondrial OS=Mus musculus OX=10090 GN=Ptrh2 PE=1 SV=1 |
|
| 2.99e-29 | 462 | 592 | 58 | 178 | Peptidyl-tRNA hydrolase 2, mitochondrial OS=Homo sapiens OX=9606 GN=PTRH2 PE=1 SV=1 |
|
| 4.10e-29 | 462 | 592 | 58 | 178 | Peptidyl-tRNA hydrolase 2, mitochondrial OS=Bos taurus OX=9913 GN=PTRH2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999982 | 0.000012 |