You are browsing environment: FUNGIDB
CAZyme Information: KAF5680204.1
Basic Information
help
| Species |
Fusarium circinatum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum
|
| CAZyme ID |
KAF5680204.1
|
| CAZy Family |
GH31 |
| CAZyme Description |
tat pathway signal sequence domain-containing protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FcircinatumNRRL25331 |
13905 |
N/A |
0 |
13905
|
|
| Gene Location |
No EC number prediction in KAF5680204.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH28 |
186 |
450 |
2.7e-33 |
0.7446153846153846 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227721
|
Pgu1 |
6.38e-05 |
183 |
314 |
223 |
338 |
Polygalacturonase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
484 |
1 |
484 |
| 0.0 |
1 |
483 |
1 |
483 |
| 0.0 |
1 |
483 |
1 |
483 |
| 0.0 |
1 |
483 |
1 |
483 |
| 0.0 |
1 |
483 |
1 |
483 |
KAF5680204.1 has no PDB hit.
KAF5680204.1 has no Swissprot hit.
This protein is predicted as OTHER
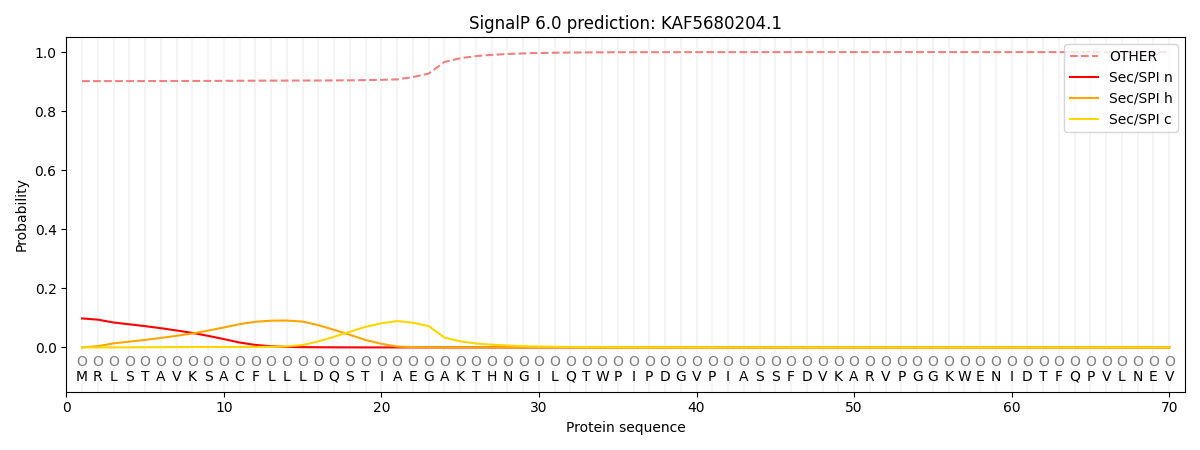
| Other |
SP_Sec_SPI |
CS Position |
| 0.906788 |
0.093241 |
|
There is no transmembrane helices in KAF5680204.1.

