You are browsing environment: FUNGIDB
CAZyme Information: KAF5678651.1
You are here: Home > Sequence: KAF5678651.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5678651.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | choline dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 41 | 601 | 3.7e-153 | 0.9947183098591549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 7.60e-67 | 38 | 602 | 4 | 536 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 3.48e-65 | 41 | 600 | 5 | 532 | choline dehydrogenase; Validated |
| 340910 | MFS_MCT_SLC16 | 1.92e-45 | 668 | 1072 | 4 | 358 | Monocarboxylate transporter (MCT) family of the Major Facilitator Superfamily of transporters. The animal Monocarboxylate transporter (MCT) family is also called Solute carrier family 16 (SLC16 or SLC16A). It is composed of 14 members, MCT1-14. MCTs play an integral role in cellular metabolism via lactate transport and have been implicated in metabolic synergy in tumors. MCT1-4 are proton-coupled transporters that facilitate the transport across the plasma membrane of monocarboxylates such as lactate, pyruvate, branched-chain oxo acids derived from leucine, valine and isoleucine, and ketone bodies such as acetoacetate, beta-hydroxybutyrate and acetate. MCT8 and MCT10 are transporters which stimulate the cellular uptake of thyroid hormones such as thyroxine (T4), triiodothyronine (T3), reverse triiodothyronine (rT3) and diidothyronine (T2). MCT10 also functions as a sodium-independent transporter that mediates the uptake or efflux of aromatic acids. Many members are orphan transporters whose substrates are yet to be determined. The MCT family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 398739 | GMC_oxred_C | 1.06e-39 | 461 | 595 | 3 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 369468 | MFS_1 | 1.55e-23 | 741 | 1028 | 66 | 345 | Major Facilitator Superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 606 | 1 | 606 | |
| 0.0 | 1 | 606 | 1 | 606 | |
| 0.0 | 1 | 606 | 1 | 606 | |
| 0.0 | 1 | 603 | 1 | 603 | |
| 0.0 | 1 | 606 | 1 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.04e-52 | 42 | 594 | 8 | 555 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
|
| 5.02e-52 | 42 | 594 | 8 | 555 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRV_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRX_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRZ_AAA Chain AAA, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
|
| 5.02e-52 | 42 | 594 | 8 | 555 | Chain A, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis],6ZH7_B Chain B, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
|
| 6.64e-52 | 42 | 594 | 24 | 571 | Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis],5NCC_B Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis],5NCC_C Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis],5NCC_D Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis],5NCC_E Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis],5NCC_F Structure of Fatty acid Photodecarboxylase in complex with FAD and palmitic acid [Chlorella variabilis] |
|
| 1.24e-51 | 42 | 594 | 8 | 555 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.22e-138 | 27 | 602 | 37 | 627 | Patulin synthase OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=patE PE=1 SV=1 |
|
| 9.87e-136 | 27 | 599 | 38 | 624 | Patulin synthase OS=Penicillium expansum OX=27334 GN=patE PE=1 SV=1 |
|
| 4.74e-134 | 3 | 595 | 6 | 603 | Dehydrogenase pkfF OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pkfF PE=2 SV=1 |
|
| 7.56e-133 | 4 | 600 | 31 | 644 | Versicolorin B synthase OS=Dothistroma septosporum (strain NZE10 / CBS 128990) OX=675120 GN=vbsA PE=2 SV=1 |
|
| 7.56e-133 | 4 | 600 | 31 | 644 | Versicolorin B synthase OS=Dothistroma septosporum OX=64363 GN=vbsA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000627 | 0.999346 | CS pos: 16-17. Pr: 0.9687 |
TMHMM Annotations download full data without filtering help
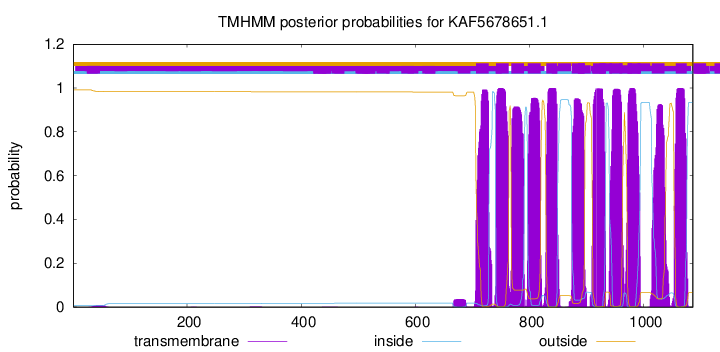
| Start | End |
|---|---|
| 706 | 728 |
| 741 | 763 |
| 768 | 790 |
| 797 | 819 |
| 829 | 848 |
| 874 | 896 |
| 911 | 933 |
| 940 | 962 |
| 972 | 994 |
| 1014 | 1036 |
| 1056 | 1075 |
