You are browsing environment: FUNGIDB
CAZyme Information: KAF5675102.1
You are here: Home > Sequence: KAF5675102.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5675102.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | pectate lyase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 80 | 253 | 1.5e-106 | 0.9943181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 2.55e-57 | 83 | 252 | 12 | 186 | Amb_all domain. |
| 226384 | PelB | 6.58e-52 | 56 | 335 | 62 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 2.54e-34 | 81 | 252 | 28 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 197609 | LysM | 1.90e-05 | 407 | 437 | 1 | 31 | Lysin motif. |
| 212030 | LysM | 7.25e-05 | 407 | 435 | 2 | 30 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 495 | 1 | 495 | |
| 0.0 | 1 | 495 | 1 | 498 | |
| 2.72e-253 | 1 | 336 | 1 | 336 | |
| 1.37e-236 | 21 | 336 | 6 | 324 | |
| 2.90e-227 | 1 | 336 | 1 | 336 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.68e-38 | 29 | 266 | 3 | 261 | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
|
| 4.71e-31 | 85 | 335 | 130 | 415 | Structure of the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
|
| 1.32e-28 | 42 | 335 | 21 | 331 | Catalytic function and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
|
| 3.92e-25 | 32 | 252 | 10 | 276 | Chain A, PECTATE LYASE E [Dickeya chrysanthemi] |
|
| 1.31e-24 | 85 | 230 | 125 | 296 | Structural insights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.05e-54 | 9 | 335 | 14 | 339 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
|
| 6.05e-54 | 9 | 335 | 14 | 339 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
|
| 6.05e-54 | 9 | 335 | 14 | 339 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
|
| 2.22e-53 | 32 | 316 | 44 | 307 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
|
| 1.16e-52 | 1 | 260 | 1 | 266 | Probable pectate lyase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
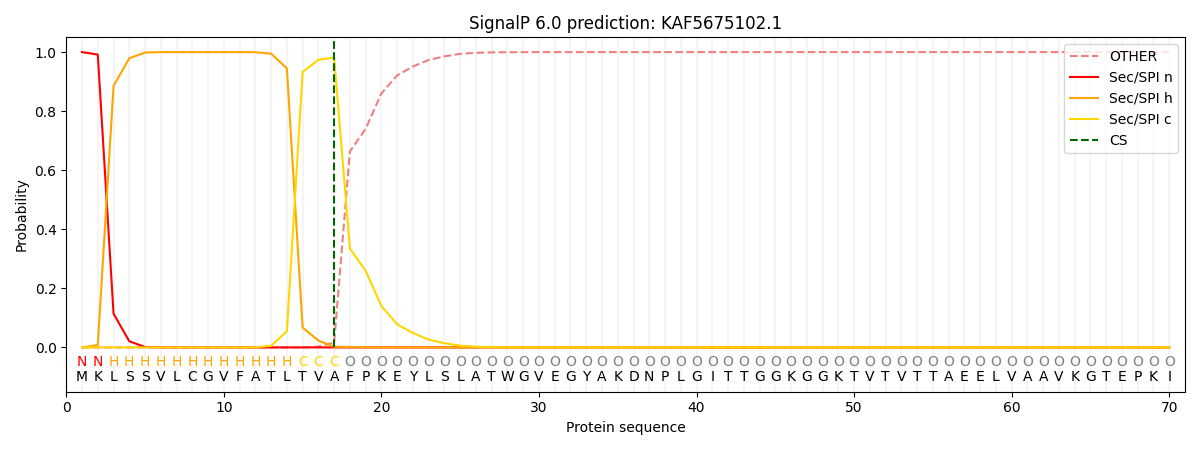
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000242 | 0.999734 | CS pos: 17-18. Pr: 0.9812 |
