You are browsing environment: FUNGIDB
CAZyme Information: KAF5670136.1
You are here: Home > Sequence: KAF5670136.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5670136.1 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | phosphoribosyl transferase domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 291 | 521 | 7.2e-41 | 0.4650655021834061 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235028 | PRK02304 | 1.00e-22 | 937 | 1118 | 12 | 175 | adenine phosphoribosyltransferase; Provisional |
| 398111 | P-mevalo_kinase | 4.61e-21 | 740 | 853 | 1 | 110 | Phosphomevalonate kinase. Phosphomevalonate kinase (EC:2.7.4.2) catalyzes the phosphorylation of 5-phosphomevalonate into 5-diphosphomevalonate, an essential step in isoprenoid biosynthesis via the mevalonate pathway. This family represents the animal type of the enzyme. The other is the ERG8 type, found in plants and fungi, and some bacteria (see pfam00288). |
| 223577 | Apt | 1.10e-18 | 924 | 1100 | 1 | 164 | Adenine/guanine phosphoribosyltransferase or related PRPP-binding protein [Nucleotide transport and metabolism]. |
| 396238 | FAD_binding_4 | 2.98e-18 | 300 | 429 | 1 | 130 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 177930 | PLN02293 | 3.41e-15 | 935 | 1100 | 21 | 173 | adenine phosphoribosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.23e-10 | 286 | 506 | 59 | 259 | |
| 5.64e-10 | 286 | 506 | 59 | 259 | |
| 5.64e-10 | 286 | 506 | 59 | 259 | |
| 7.21e-10 | 286 | 720 | 53 | 482 | |
| 9.54e-10 | 286 | 720 | 53 | 483 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.63e-17 | 925 | 1118 | 3 | 180 | Crystal structure of an APRT from Yersinia pseudotuberculosis in complex with AMP. [Yersinia pseudotuberculosis IP 32953] |
|
| 5.34e-17 | 925 | 1118 | 9 | 186 | Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis. [Yersinia pseudotuberculosis IP 32953],5Y07_A Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis with PRPP. [Yersinia pseudotuberculosis IP 32953],5Y07_B Crystal structure of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis with PRPP. [Yersinia pseudotuberculosis IP 32953],5Y4A_A Cadmium directed assembly of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis. [Yersinia pseudotuberculosis IP 32953],5Y4A_B Cadmium directed assembly of adenine phosphoribosyltransferase from Yersinia pseudotuberculosis. [Yersinia pseudotuberculosis IP 32953],5ZC7_A Crystal structure of APRT from Y. pseudotuberculosis with bound adenine (P63 space group). [Yersinia pseudotuberculosis IP 32953],5ZC7_B Crystal structure of APRT from Y. pseudotuberculosis with bound adenine (P63 space group). [Yersinia pseudotuberculosis IP 32953],5ZMI_A Crystal structure of APRT from Y. pseudotuberculosis in complex with adenine. [Yersinia pseudotuberculosis IP 32953],5ZNQ_A Crystal structure of APRT from Y. pseudotuberculosis with bound adenine (P21 space group). [Yersinia pseudotuberculosis IP 32953],5ZNQ_B Crystal structure of APRT from Y. pseudotuberculosis with bound adenine (P21 space group). [Yersinia pseudotuberculosis IP 32953],5ZOC_A Crystal structure of APRT from Y. pseudotuberculosis with bound adenine (C2 space group). [Yersinia pseudotuberculosis IP 32953] |
|
| 5.10e-16 | 300 | 476 | 39 | 198 | Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 3 (P1) [Paenarthrobacter nicotinovorans],2BVF_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 3 (P1) [Paenarthrobacter nicotinovorans],2BVG_A Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_C Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVG_D Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 1 (P21) [Paenarthrobacter nicotinovorans],2BVH_A Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_B Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_C Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans],2BVH_D Crystal structure of 6-hydoxy-D-nicotine oxidase from Arthrobacter nicotinovorans. Crystal Form 2 (P21) [Paenarthrobacter nicotinovorans] |
|
| 1.01e-15 | 923 | 1100 | 20 | 184 | Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. [Thermoanaerobacter pseudethanolicus ATCC 33223],4LZA_B Crystal structure of adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus ATCC 33223, NYSGRC Target 029700. [Thermoanaerobacter pseudethanolicus ATCC 33223] |
|
| 3.57e-14 | 925 | 1118 | 12 | 189 | Crystal structure of project JW0458 from Escherichia coli [Escherichia coli K-12],2DY0_B Crystal structure of project JW0458 from Escherichia coli [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.11e-31 | 298 | 719 | 46 | 439 | FAD-linked oxidoreductase DDB_G0289697 OS=Dictyostelium discoideum OX=44689 GN=DDB_G0289697 PE=2 SV=1 |
|
| 2.17e-19 | 930 | 1105 | 4 | 166 | Adenine phosphoribosyltransferase OS=Roseiflexus sp. (strain RS-1) OX=357808 GN=apt PE=3 SV=1 |
|
| 2.95e-19 | 929 | 1105 | 3 | 166 | Adenine phosphoribosyltransferase OS=Roseiflexus castenholzii (strain DSM 13941 / HLO8) OX=383372 GN=apt PE=3 SV=1 |
|
| 1.78e-18 | 929 | 1100 | 1 | 159 | Adenine phosphoribosyltransferase OS=Kosmotoga olearia (strain ATCC BAA-1733 / DSM 21960 / TBF 19.5.1) OX=521045 GN=apt PE=3 SV=1 |
|
| 1.18e-17 | 924 | 1119 | 4 | 184 | Adenine phosphoribosyltransferase OS=Shewanella putrefaciens (strain CN-32 / ATCC BAA-453) OX=319224 GN=apt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
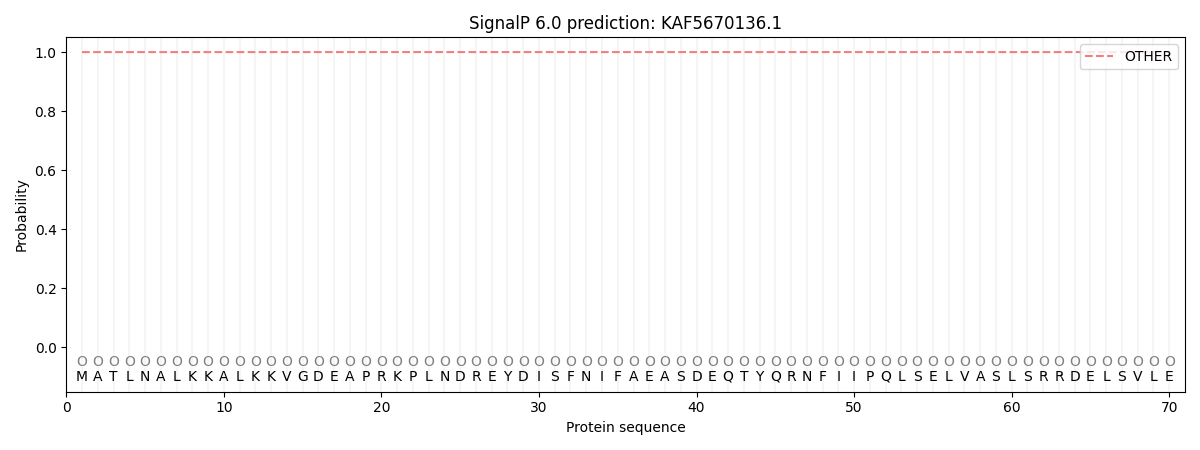
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000063 | 0.000000 |
