You are browsing environment: FUNGIDB
CAZyme Information: KAF5666909.1
You are here: Home > Sequence: KAF5666909.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5666909.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | SIK1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:25 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 906 | 1259 | 1.9e-78 | 0.9594594594594594 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119365 | GH18_chitinase | 1.33e-127 | 908 | 1251 | 2 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| 224415 | SIK1 | 7.05e-125 | 1357 | 1734 | 2 | 363 | RNA processing factor Prp31, contains Nop domain [Translation, ribosomal structure and biogenesis]. |
| 214753 | Glyco_18 | 8.31e-119 | 906 | 1251 | 1 | 334 | Glyco_18 domain. |
| 340885 | MFS_FEN2_like | 7.44e-117 | 393 | 799 | 1 | 406 | Pantothenate transporter FEN2 and similar transporters of the Major Facilitator Superfamily. This family is composed of Saccharomyces cerevisiae pantothenate transporter FEN2 (or fenpropimorph resistance protein 2) and similar proteins from fungi and bacteria including fungal vitamin H transporter, allantoate permease, and high-affinity nicotinic acid transporter, as well as Pseudomonas putida phthalate transporter and nicotinate degradation protein T (nicT). These proteins are involved in the uptake into the cell of specific substrates such as pathothenate, biotin, allantoate, and nicotinic acid, among others. The FEN2-like family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 396390 | Nop | 8.13e-108 | 1496 | 1724 | 5 | 228 | snoRNA binding domain, fibrillarin. This family consists of various Pre RNA processing ribonucleoproteins. The function of the aligned region is unknown however it may be a common RNA or snoRNA or Nop1p binding domain. Nop5p (Nop58p) from yeast is the protein component of a ribonucleoprotein required for pre-18s rRNA processing and is suggested to function with Nop1p in a snoRNA complex. Nop56p and Nop5p interact with Nop1p and are required for ribosome biogenesis. Prp31p is required for pre-mRNA splicing in S. cerevisiae. Fibrillarin, or Nop, is the catalytic subunit responsible for the methyl transfer reaction of the site-specific 2'-O-methylation of ribosomal and spliceosomal RNA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 898 | 1769 | 1 | 851 | |
| 0.0 | 898 | 1769 | 1 | 837 | |
| 0.0 | 898 | 1769 | 1 | 837 | |
| 0.0 | 898 | 1769 | 1 | 837 | |
| 0.0 | 898 | 1767 | 1 | 847 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.18e-184 | 1312 | 1740 | 4 | 432 | Chain T, Putative nucleolar protein [Thermochaetoides thermophila DSM 1495],6RXT_CC Chain CC, Putative nucleolar protein [Thermochaetoides thermophila],6RXU_CC Chain CC, Nop56 [Thermochaetoides thermophila],6RXV_CC Chain CC, Nop56 [Thermochaetoides thermophila DSM 1495],6RXX_CC Chain CC, Putative nucleolar protein [Thermochaetoides thermophila],6RXY_CC Chain CC, Nop56 [Thermochaetoides thermophila],6RXZ_CC Chain CC, Putative nucleolar protein [Thermochaetoides thermophila] |
|
| 1.14e-169 | 1309 | 1769 | 1 | 452 | Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],5WYK_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6KE6_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQP_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQQ_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQR_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQS_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQT_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQU_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6LQV_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6ZQA_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6ZQB_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6ZQC_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6ZQD_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],6ZQE_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7AJT_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7AJU_CD Chain CD, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7D4I_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7D5S_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7D5T_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C],7D63_3D Chain 3D, Nucleolar protein 56 [Saccharomyces cerevisiae S288C] |
|
| 6.87e-156 | 904 | 1287 | 4 | 387 | Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 [Aspergillus fumigatus],1WNO_B Crystal structure of a native chitinase from Aspergillus fumigatus YJ-407 [Aspergillus fumigatus] |
|
| 9.74e-156 | 884 | 1287 | 21 | 425 | Specificity and affinity of natural product cyclopentapeptide inhibitors against Aspergillus fumigatus, human and bacterial chitinaseFra [Aspergillus fumigatus],1W9P_B Specificity and affinity of natural product cyclopentapeptide inhibitors against Aspergillus fumigatus, human and bacterial chitinaseFra [Aspergillus fumigatus],1W9U_A Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase [Aspergillus fumigatus],1W9U_B Specificity and affnity of natural product cyclopentapeptide inhibitor Argadin against Aspergillus fumigatus chitinase [Aspergillus fumigatus],1W9V_A Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus [Aspergillus fumigatus],1W9V_B Specificity and affinity of natural product cyclopentapeptide argifin against Aspergillus fumigatus [Aspergillus fumigatus],2A3A_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline [Aspergillus fumigatus],2A3A_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with theophylline [Aspergillus fumigatus],2A3B_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with caffeine [Aspergillus fumigatus],2A3B_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with caffeine [Aspergillus fumigatus],2A3C_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with pentoxifylline [Aspergillus fumigatus],2A3C_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with pentoxifylline [Aspergillus fumigatus],2A3E_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin [Aspergillus fumigatus],2A3E_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with allosamidin [Aspergillus fumigatus],2IUZ_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with C2-dicaffeine [Aspergillus fumigatus],2IUZ_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with C2-dicaffeine [Aspergillus fumigatus],3CH9_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dimethylguanylurea [Aspergillus fumigatus],3CH9_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dimethylguanylurea [Aspergillus fumigatus],3CHC_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with monopeptide [Aspergillus fumigatus],3CHC_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with monopeptide [Aspergillus fumigatus],3CHD_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dipeptide [Aspergillus fumigatus],3CHD_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with dipeptide [Aspergillus fumigatus],3CHE_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tripeptide [Aspergillus fumigatus],3CHE_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tripeptide [Aspergillus fumigatus],3CHF_A Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tetrapeptide [Aspergillus fumigatus],3CHF_B Crystal structure of Aspergillus fumigatus chitinase B1 in complex with tetrapeptide [Aspergillus fumigatus] |
|
| 2.24e-155 | 905 | 1287 | 16 | 397 | Crystal structure of Aspergillus niger chitinase B [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.87e-169 | 1309 | 1769 | 1 | 452 | Nucleolar protein 56 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=NOP56 PE=1 SV=1 |
|
| 3.44e-162 | 1313 | 1758 | 3 | 443 | Nucleolar protein 56 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=nop56 PE=3 SV=1 |
|
| 5.01e-155 | 884 | 1287 | 21 | 425 | Endochitinase B1 OS=Neosartorya fumigata OX=746128 GN=chiB1 PE=1 SV=1 |
|
| 5.01e-155 | 884 | 1287 | 21 | 425 | Endochitinase B1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chiB1 PE=3 SV=1 |
|
| 1.88e-148 | 892 | 1282 | 25 | 414 | Endochitinase 1 OS=Coccidioides posadasii (strain RMSCC 757 / Silveira) OX=443226 GN=CTS1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000078 | 0.000000 |
TMHMM Annotations download full data without filtering help
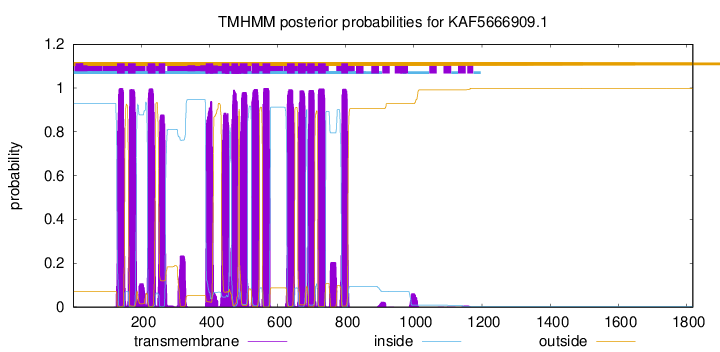
| Start | End |
|---|---|
| 129 | 151 |
| 161 | 183 |
| 219 | 241 |
| 251 | 270 |
| 389 | 411 |
| 436 | 458 |
| 465 | 484 |
| 489 | 511 |
| 524 | 546 |
| 556 | 578 |
| 628 | 650 |
| 660 | 682 |
| 687 | 709 |
| 719 | 741 |
| 787 | 806 |
