You are browsing environment: FUNGIDB
CAZyme Information: KAF5664599.1
You are here: Home > Sequence: KAF5664599.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5664599.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403281 | DUF3500 | 2.80e-162 | 82 | 405 | 1 | 292 | Protein of unknown function (DUF3500). This family of proteins is functionally uncharacterized. This protein is found in bacteria and eukaryotes. Proteins in this family are typically between 335 to 438 amino acids in length. This protein has a conserved GHH sequence motif. This protein has two completely conserved G residues that may be functionally important. |
| 404972 | RicinB_lectin_2 | 7.89e-11 | 421 | 509 | 9 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.59e-118 | 414 | 580 | 1 | 167 | |
| 1.11e-103 | 414 | 580 | 1 | 152 | |
| 1.11e-103 | 414 | 580 | 1 | 152 | |
| 1.11e-103 | 414 | 580 | 1 | 152 | |
| 1.11e-103 | 414 | 580 | 1 | 152 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.27e-16 | 431 | 575 | 12 | 151 | MOA-E-64 complex [Marasmius oreades] |
|
| 2.31e-16 | 431 | 575 | 12 | 151 | Crystal structure of MOA, a lectin from the mushroom Marasmius oreades in complex with the trisaccharide Gal(1,3)Gal(1,4)GlcNAc [Marasmius oreades],5D61_A MOA-Z-VAD-fmk complex, direct orientation [Marasmius oreades],5D62_A MOA-Z-VAD-fmk complex, inverted orientation [Marasmius oreades],5D63_A MOA-Z-VAD-fmk inhibitor complex, direct/inverted dual orientation [Marasmius oreades],6TSO_AAA Chain AAA, Agglutinin [Marasmius oreades],6TSP_AAA Chain AAA, Agglutinin [Marasmius oreades] |
|
| 2.31e-16 | 431 | 575 | 12 | 151 | Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. [Marasmius oreades],3EF2_B Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. [Marasmius oreades],3EF2_C Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. [Marasmius oreades],3EF2_D Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. [Marasmius oreades],6TSQ_AAA Chain AAA, Agglutinin [Marasmius oreades],6TSR_AAA Chain AAA, Agglutinin [Marasmius oreades] |
|
| 2.31e-16 | 431 | 575 | 12 | 151 | Chain AAA, Agglutinin [Marasmius oreades] |
|
| 2.31e-16 | 431 | 575 | 12 | 151 | Chain AAA, Agglutinin [Marasmius oreades] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
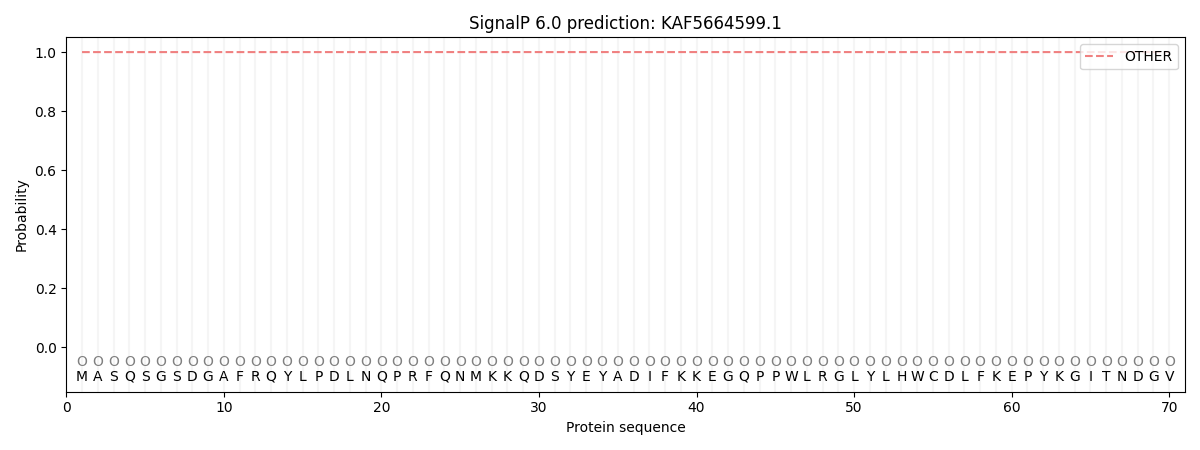
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000000 |
