You are browsing environment: FUNGIDB
CAZyme Information: KAF5661968.1
Basic Information
help
| Species |
Fusarium circinatum
|
| Lineage |
Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum
|
| CAZyme ID |
KAF5661968.1
|
| CAZy Family |
AA7 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_FcircinatumNRRL25331 |
13905 |
N/A |
0 |
13905
|
|
| Gene Location |
No EC number prediction in KAF5661968.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH12 |
136 |
280 |
3e-20 |
0.8589743589743589 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396303
|
Glyco_hydro_12 |
1.77e-14 |
43 |
281 |
8 |
206 |
Glycosyl hydrolase family 12. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
41 |
701 |
174 |
851 |
| 0.0 |
41 |
701 |
174 |
851 |
| 0.0 |
41 |
701 |
174 |
851 |
| 0.0 |
38 |
655 |
109 |
772 |
| 9.74e-215 |
1 |
344 |
1 |
341 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.40e-15 |
12 |
277 |
5 |
230 |
CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550],5M2D_B CRYSTAL STRUCTURE 4Ac Endoglucanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.18e-31 |
345 |
701 |
443 |
800 |
FAD-dependent monooxygenase verC1 OS=Penicillium polonicum OX=60169 GN=verC1 PE=3 SV=1 |
| 1.06e-14 |
430 |
691 |
494 |
755 |
FAD-dependent monooxygenase ntnK OS=Nectria sp. OX=1755444 GN=ntnK PE=2 SV=1 |
| 1.27e-10 |
446 |
689 |
524 |
771 |
FAD-dependent monooxygenase BOA8 OS=Botryotinia fuckeliana (strain B05.10) OX=332648 GN=BOA8 PE=2 SV=1 |
| 6.28e-06 |
428 |
657 |
80 |
336 |
Terpene cyclase ascI OS=Acremonium egyptiacum OX=749675 GN=ascI PE=1 SV=1 |
This protein is predicted as SP
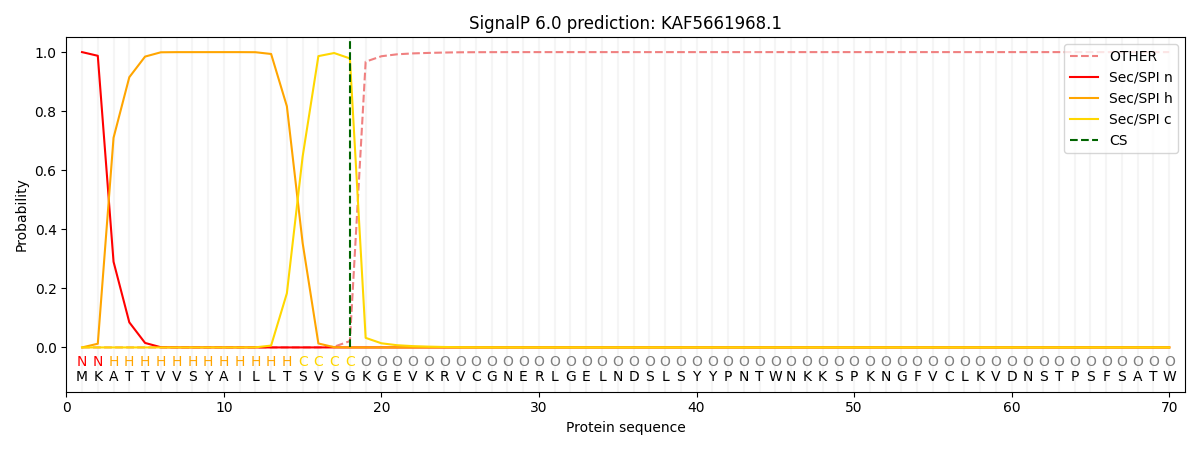
| Other |
SP_Sec_SPI |
CS Position |
| 0.000551 |
0.999430 |
CS pos: 18-19. Pr: 0.9785 |
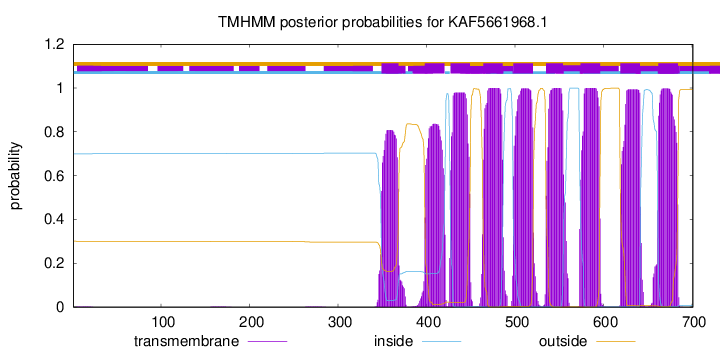
| Start |
End |
| 349 |
368 |
| 398 |
420 |
| 427 |
449 |
| 464 |
486 |
| 498 |
520 |
| 535 |
554 |
| 574 |
596 |
| 619 |
641 |
| 662 |
684 |


