You are browsing environment: FUNGIDB
CAZyme Information: KAF5659773.1
You are here: Home > Sequence: KAF5659773.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5659773.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | sterigmatocystin biosynthesis peroxidase stcC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396065 | Peroxidase_2 | 7.26e-52 | 424 | 619 | 1 | 177 | Peroxidase, family 2. The peroxidases in this family do not have similarity to other peroxidases. |
| 340817 | GT1_Gtf-like | 6.11e-12 | 39 | 407 | 3 | 354 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 224732 | YjiC | 7.12e-05 | 36 | 418 | 1 | 358 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-256 | 1 | 408 | 1 | 450 | |
| 6.42e-157 | 7 | 407 | 5 | 450 | |
| 5.68e-156 | 1 | 407 | 1 | 450 | |
| 8.23e-155 | 6 | 407 | 4 | 450 | |
| 8.23e-155 | 6 | 407 | 4 | 450 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.04e-29 | 420 | 652 | 3 | 226 | Chain A, Marasmius rotula UPO [Marasmius rotula],7ZBP_B Chain B, Marasmius rotula UPO [Marasmius rotula],7ZBP_C Chain C, Marasmius rotula UPO [Marasmius rotula],7ZBP_D Chain D, Marasmius rotula UPO [Marasmius rotula] |
|
| 1.26e-29 | 424 | 652 | 3 | 222 | Crystallization of a dimeric heme peroxygenase from the fungus Marasmius rotula [Marasmius rotula],5FUK_B Crystallization of a dimeric heme peroxygenase from the fungus Marasmius rotula [Marasmius rotula] |
|
| 1.32e-29 | 424 | 652 | 3 | 222 | Crystallization of a dimeric heme peroxygenase from the fungus Marasmius rotula [Marasmius rotula],5FUJ_B Crystallization of a dimeric heme peroxygenase from the fungus Marasmius rotula [Marasmius rotula] |
|
| 1.28e-26 | 425 | 625 | 5 | 197 | Chain A, Collariella virescens UPO [Achaetomiella virescens] |
|
| 1.31e-26 | 425 | 625 | 6 | 198 | Chain B, Collariella virescens UPO [Achaetomiella virescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.82e-54 | 34 | 429 | 3 | 433 | Glycosyltransferase buaB OS=Aspergillus burnettii OX=2508778 GN=buaB PE=3 SV=1 |
|
| 2.20e-53 | 80 | 409 | 99 | 433 | Glycosyltransferase sdnJ OS=Sordaria araneosa OX=573841 GN=sdnJ PE=1 SV=1 |
|
| 9.23e-16 | 405 | 627 | 12 | 225 | Peroxidase stcC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=stcC PE=2 SV=2 |
|
| 1.36e-10 | 424 | 535 | 22 | 146 | Aromatic peroxygenase (Fragments) OS=Coprinellus radians OX=71721 GN=APO PE=1 SV=2 |
|
| 2.47e-07 | 428 | 534 | 40 | 156 | Chloroperoxidase OS=Leptoxyphium fumago OX=5474 GN=CPO PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
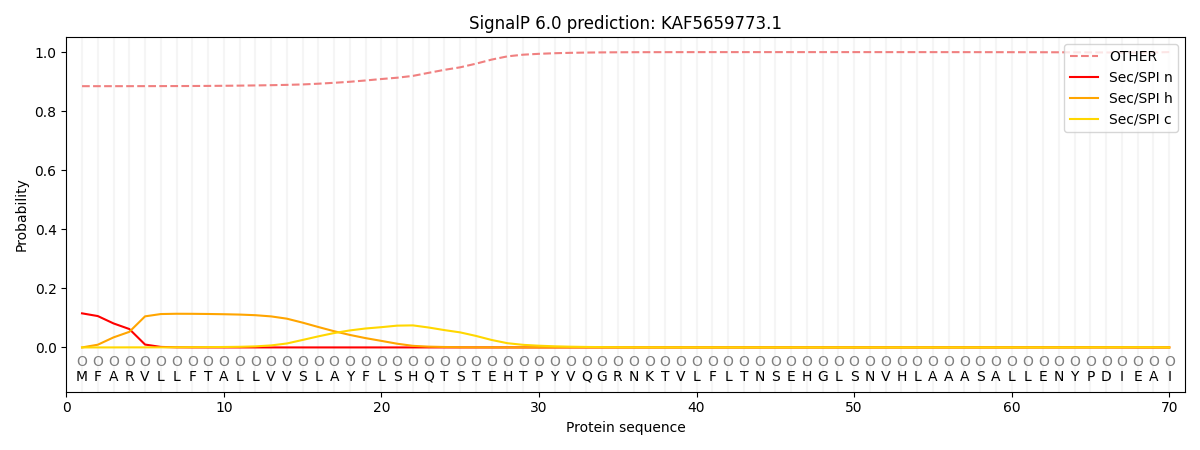
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.891390 | 0.108639 |
