You are browsing environment: FUNGIDB
CAZyme Information: KAF5658606.1
You are here: Home > Sequence: KAF5658606.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5658606.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | lactose permease | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 560 | 1017 | 1.5e-140 | 0.951048951048951 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 1.72e-145 | 560 | 1015 | 2 | 431 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 2.49e-119 | 562 | 1021 | 3 | 443 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215455 | PLN02849 | 5.67e-95 | 560 | 1028 | 27 | 486 | beta-glucosidase |
| 215435 | PLN02814 | 9.21e-94 | 556 | 1013 | 21 | 460 | beta-glucosidase |
| 215539 | PLN02998 | 1.28e-86 | 560 | 1014 | 28 | 466 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 550 | 1021 | 1 | 472 | |
| 0.0 | 550 | 1021 | 1 | 472 | |
| 0.0 | 550 | 1021 | 1 | 472 | |
| 0.0 | 550 | 1021 | 1 | 472 | |
| 0.0 | 550 | 1021 | 1 | 472 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.45e-153 | 563 | 1018 | 26 | 478 | Chain A, Beta-glucosidase [[Humicola] grisea var. thermoidea],4MDP_A Chain A, Beta-glucosidase [[Humicola] grisea var. thermoidea] |
|
| 3.27e-151 | 562 | 1019 | 20 | 476 | Chain A, Glycoside hydrolase family 1 [Trichoderma reesei QM6a],6KHT_B Chain B, Glycoside hydrolase family 1 [Trichoderma reesei QM6a],6KHT_C Chain C, Glycoside hydrolase family 1 [Trichoderma reesei QM6a] |
|
| 1.30e-150 | 563 | 1015 | 3 | 442 | Crystal structure of the double mutant L167W / P172L of the beta-glucosidase from Trichoderma harzianum [Trichoderma harzianum],6EFU_B Crystal structure of the double mutant L167W / P172L of the beta-glucosidase from Trichoderma harzianum [Trichoderma harzianum] |
|
| 2.52e-150 | 563 | 1015 | 3 | 442 | Trichoderma harzianum GH1 beta-glucosidase ThBgl1 [Trichoderma harzianum],5JBK_B Trichoderma harzianum GH1 beta-glucosidase ThBgl1 [Trichoderma harzianum] |
|
| 4.81e-150 | 563 | 1015 | 22 | 461 | Crystal structure of the beta-glucosidase from Trichoderma harzianum [Trichoderma harzianum],5BWF_B Crystal structure of the beta-glucosidase from Trichoderma harzianum [Trichoderma harzianum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-142 | 563 | 1025 | 11 | 465 | Beta-glucosidase 1B OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1B PE=1 SV=1 |
|
| 1.04e-135 | 563 | 1019 | 6 | 444 | Beta-glucosidase 1A OS=Phanerodontia chrysosporium OX=2822231 GN=BGL1A PE=1 SV=1 |
|
| 1.81e-118 | 560 | 1016 | 93 | 544 | Furostanol glycoside 26-O-beta-glucosidase OS=Hellenia speciosa OX=49577 GN=F26G PE=1 SV=1 |
|
| 1.26e-117 | 560 | 1018 | 47 | 505 | Beta-glucosidase 6 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU6 PE=1 SV=1 |
|
| 1.15e-116 | 560 | 1018 | 34 | 492 | Beta-glucosidase 40 OS=Arabidopsis thaliana OX=3702 GN=BGLU40 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999796 | 0.000221 |
TMHMM Annotations download full data without filtering help
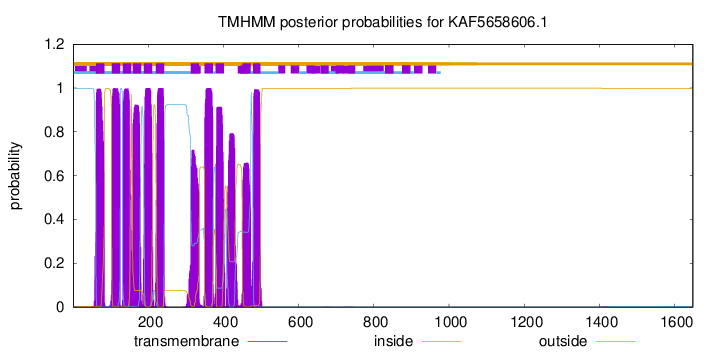
| Start | End |
|---|---|
| 61 | 83 |
| 103 | 125 |
| 132 | 154 |
| 159 | 181 |
| 188 | 210 |
| 220 | 242 |
| 313 | 335 |
| 350 | 372 |
| 379 | 401 |
| 450 | 472 |
| 479 | 498 |
