You are browsing environment: FUNGIDB
CAZyme Information: KAF5656113.1
You are here: Home > Sequence: KAF5656113.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5656113.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | cellulose hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 189 | 387 | 6.7e-69 | 0.44243792325056436 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227807 | XynC | 1.27e-11 | 36 | 387 | 35 | 427 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 407314 | Glyco_hydro_30C | 1.13e-05 | 296 | 386 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
| 405300 | Glyco_hydr_30_2 | 7.97e-04 | 71 | 187 | 115 | 232 | O-Glycosyl hydrolase family 30. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.10e-239 | 1 | 390 | 1 | 485 | |
| 8.95e-236 | 1 | 390 | 1 | 485 | |
| 8.95e-236 | 1 | 390 | 1 | 485 | |
| 8.95e-236 | 1 | 390 | 1 | 485 | |
| 8.95e-236 | 1 | 390 | 1 | 485 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.26e-132 | 37 | 388 | 24 | 471 | Chain A, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
|
| 1.05e-131 | 34 | 388 | 92 | 542 | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
|
| 1.15e-77 | 37 | 387 | 5 | 446 | Chain A, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464],7O0E_G Chain G, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.97e-76 | 37 | 387 | 12 | 453 | Chain AAA, GH30 family xylanase [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.22e-61 | 37 | 387 | 25 | 452 | Chain A, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.47e-77 | 37 | 387 | 29 | 470 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
|
| 4.94e-07 | 113 | 387 | 145 | 420 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
|
| 4.77e-06 | 113 | 387 | 144 | 419 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
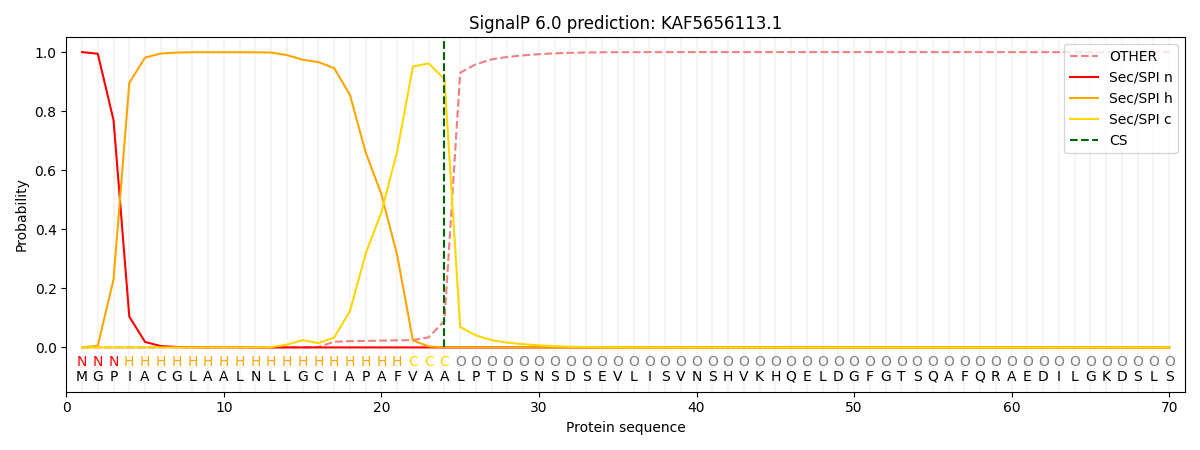
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000726 | 0.999251 | CS pos: 24-25. Pr: 0.9088 |
