You are browsing environment: FUNGIDB
CAZyme Information: KAF5655914.1
You are here: Home > Sequence: KAF5655914.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusarium circinatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Nectriaceae; Fusarium; Fusarium circinatum | |||||||||||
| CAZyme ID | KAF5655914.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | alpha-n-acetylgalactosaminidase precursor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.22:1 | 3.2.1.88:1 | 3.2.1.22:1 | 3.2.1.88:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 131 | 379 | 1.2e-59 | 0.9737991266375546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 269893 | GH27 | 1.15e-99 | 26 | 315 | 1 | 271 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 178295 | PLN02692 | 4.47e-69 | 25 | 379 | 55 | 385 | alpha-galactosidase |
| 166449 | PLN02808 | 1.83e-66 | 25 | 393 | 31 | 376 | alpha-galactosidase |
| 177874 | PLN02229 | 7.91e-66 | 25 | 393 | 62 | 410 | alpha-galactosidase |
| 374582 | Melibiase_2 | 2.79e-48 | 25 | 315 | 1 | 284 | Alpha galactosidase A. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 549 | |
| 0.0 | 1 | 549 | 1 | 551 | |
| 0.0 | 1 | 549 | 1 | 551 | |
| 0.0 | 1 | 549 | 1 | 551 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.50e-56 | 25 | 379 | 8 | 338 | Nicotiana benthamiana alpha-galactosidase [Nicotiana benthamiana] |
|
| 7.78e-56 | 25 | 393 | 8 | 352 | Chain A, alpha-galactosidase [Oryza sativa] |
|
| 2.80e-52 | 25 | 394 | 8 | 384 | Crystal structure of alpha-galactosidase I from Mortierella vinacea [Umbelopsis vinacea] |
|
| 8.69e-50 | 26 | 398 | 12 | 408 | Chain A, alpha-galactosidase [Trichoderma reesei],1T0O_A Chain A, alpha-galactosidase [Trichoderma reesei] |
|
| 3.68e-44 | 25 | 385 | 29 | 407 | Chain A, Alpha-galactosidase 1 [Saccharomyces cerevisiae],3LRL_A Chain A, Alpha-galactosidase 1 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.94e-72 | 7 | 402 | 15 | 403 | Probable alpha-galactosidase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=aglD PE=3 SV=2 |
|
| 3.11e-70 | 25 | 400 | 30 | 400 | Probable alpha-galactosidase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=aglD PE=3 SV=2 |
|
| 3.11e-70 | 25 | 400 | 30 | 400 | Probable alpha-galactosidase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=aglD PE=3 SV=2 |
|
| 2.28e-69 | 5 | 400 | 12 | 400 | Probable alpha-galactosidase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=aglD PE=3 SV=1 |
|
| 3.77e-65 | 25 | 400 | 33 | 397 | Probable alpha-galactosidase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aglD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
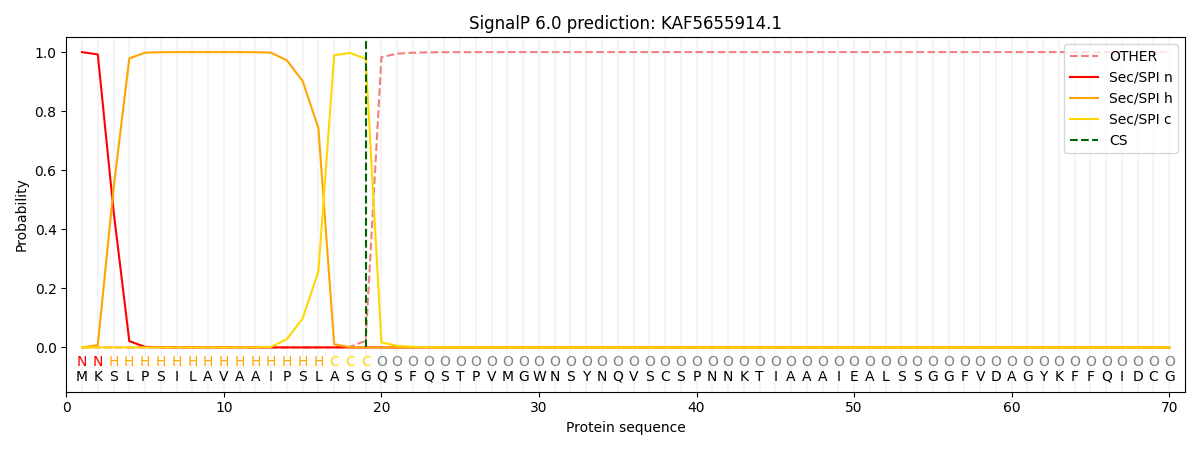
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000229 | 0.999733 | CS pos: 19-20. Pr: 0.9775 |
