You are browsing environment: FUNGIDB
CAZyme Information: KAE8541469.1
You are here: Home > Sequence: KAE8541469.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
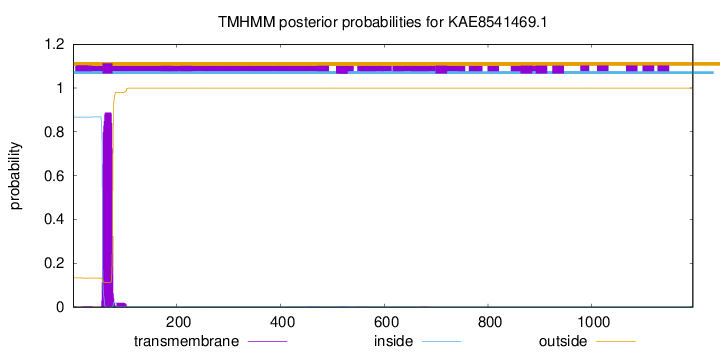
TMHMM annotations
Basic Information help
| Species | Cryptococcus cf. gattii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus cf. gattii | |||||||||||
| CAZyme ID | KAE8541469.1 | |||||||||||
| CAZy Family | GH79 | |||||||||||
| CAZyme Description | imidazoleglycerol phosphate synthase, cyclase subunit | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178226 | PLN02617 | 0.0 | 597 | 1188 | 5 | 537 | imidazole glycerol phosphate synthase hisHF |
| 240082 | HisF | 1.99e-97 | 861 | 1183 | 2 | 243 | The cyclase subunit of imidazoleglycerol phosphate synthase (HisF). Imidazole glycerol phosphate synthase (IGPS) catalyzes the fifth step of histidine biosynthesis, the formation of the imidazole ring. IGPS converts N1-(5'-phosphoribulosyl)-formimino-5-aminoimidazole-4-carboxamide ribonucleotide (PRFAR) to imidazole glycerol phosphate (ImGP) and 5'-(5-aminoimidazole-4-carboxamide) ribonucleotide (AICAR). This conversion involves two tightly coupled reactions in distinct active sites of IGPS. The two catalytic domains can be fused, like in fungi and plants, or peformed by a heterodimer (HisH-glutaminase and HisF-cyclase), like in bacteria. |
| 223185 | HisF | 1.54e-92 | 857 | 1188 | 1 | 254 | Imidazole glycerol phosphate synthase subunit HisF [Amino acid transport and metabolism]. |
| 234996 | PRK02083 | 1.81e-79 | 858 | 1188 | 2 | 252 | imidazole glycerol phosphate synthase subunit HisF; Provisional |
| 153219 | GATase1_IGP_Synthase | 7.06e-74 | 602 | 820 | 2 | 198 | Type 1 glutamine amidotransferase (GATase1) domain found in imidazole glycerol phosphate synthase (IGPS). Type 1 glutamine amidotransferase (GATase1) domain found in imidazole glycerol phosphate synthase (IGPS). IGPS incorporates ammonia derived from glutamine into N1-[(5'-phosphoribulosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (PRFAR) to form 5'-(5-aminoimidazole-4-carboxamide) ribonucleotide (AICAR) and imidazole glycerol phosphate (IGP). The glutamine amidotransferase domain generates the ammonia nucleophile which is channeled from the glutaminase active site to the PRFAR active site. IGPS belong to the triad family of amidotransferases having a conserved Cys-His-Glu catalytic triad in the glutaminase active site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 574 | 1 | 554 | |
| 0.0 | 2 | 574 | 3 | 555 | |
| 0.0 | 2 | 574 | 3 | 555 | |
| 0.0 | 2 | 581 | 3 | 562 | |
| 0.0 | 2 | 581 | 3 | 562 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-166 | 599 | 1188 | 5 | 551 | Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae],1OX6_A Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae],1OX6_B Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae] |
|
| 3.08e-165 | 599 | 1188 | 5 | 551 | Crystal Structure Of Imidazole Glycerol Phosphate Synthase: A Tunnel Through A (BetaALPHA)8 BARREL JOINS TWO ACTIVE SITES [Saccharomyces cerevisiae],1JVN_B Crystal Structure Of Imidazole Glycerol Phosphate Synthase: A Tunnel Through A (BetaALPHA)8 BARREL JOINS TWO ACTIVE SITES [Saccharomyces cerevisiae],1OX4_B Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae],1OX5_A Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae],1OX5_B Towards Understanding The Mechanism Of The Complex Cyclization Reaction Catalyzed By Imidazole Glycerophosphate Synthase [Saccharomyces cerevisiae] |
|
| 3.27e-48 | 858 | 1188 | 2 | 250 | Structure of HisF-LUCA,4EVZ_B Structure of HisF-LUCA |
|
| 8.40e-43 | 858 | 1188 | 3 | 250 | Imidazole Glycerol Phosphate Synthase [Thermus thermophilus] |
|
| 1.19e-40 | 861 | 1188 | 6 | 252 | HisF protein from Pyrobaculum aerophilum [Pyrobaculum aerophilum],1H5Y_B HisF protein from Pyrobaculum aerophilum [Pyrobaculum aerophilum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.50e-188 | 603 | 1188 | 5 | 540 | Imidazole glycerol phosphate synthase hisHF OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=his4 PE=3 SV=3 |
|
| 5.55e-184 | 599 | 1189 | 2 | 548 | Imidazole glycerol phosphate synthase hisHF OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=hisHF PE=3 SV=2 |
|
| 4.76e-166 | 599 | 1188 | 2 | 548 | Imidazole glycerol phosphate synthase hisHF OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HIS7 PE=1 SV=2 |
|
| 1.79e-165 | 603 | 1188 | 66 | 591 | Imidazole glycerol phosphate synthase hisHF, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=HISN4 PE=2 SV=1 |
|
| 1.97e-54 | 858 | 1188 | 3 | 270 | Imidazole glycerol phosphate synthase subunit HisF OS=Haloarcula marismortui (strain ATCC 43049 / DSM 3752 / JCM 8966 / VKM B-1809) OX=272569 GN=hisF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
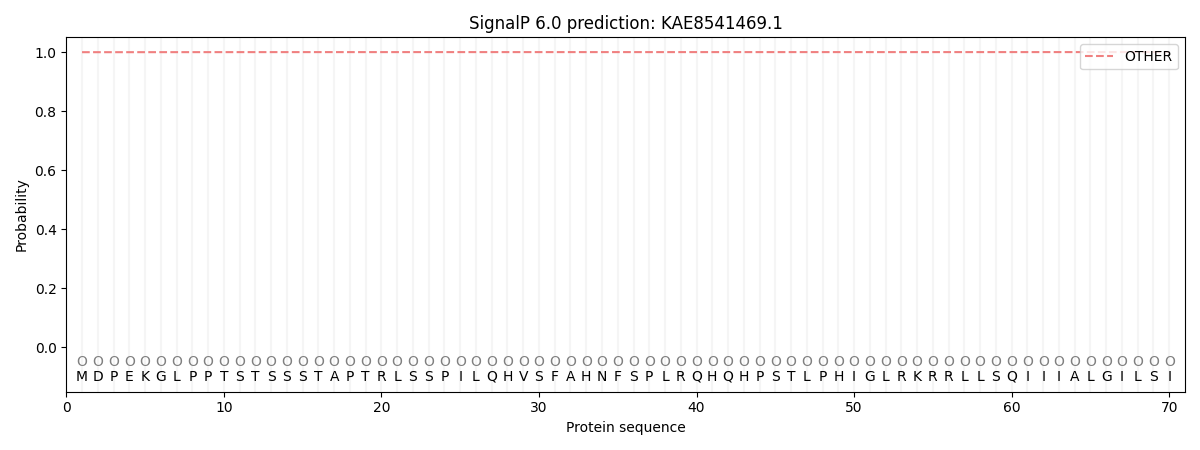
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999818 | 0.000189 |