You are browsing environment: FUNGIDB
CAZyme Information: KAE8541097.1
You are here: Home > Sequence: KAE8541097.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cryptococcus cf. gattii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus cf. gattii | |||||||||||
| CAZyme ID | KAE8541097.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 144 | 412 | 4.4e-77 | 0.9961832061068703 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 252941 | Mannosyl_trans | 8.03e-56 | 144 | 412 | 1 | 259 | Mannosyltransferase (PIG-M). PIG-M has a DXD motif. The DXD motif is found in many glycosyltransferases that utilize nucleotide sugars. It is thought that the motif is involved in the binding of a manganese ion that is required for association of the enzymes with nucleotide sugar substrates. |
| 178434 | PLN02841 | 4.75e-53 | 9 | 412 | 11 | 415 | GPI mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.18e-295 | 1 | 427 | 1 | 423 | |
| 1.10e-293 | 1 | 425 | 1 | 421 | |
| 9.34e-274 | 1 | 426 | 1 | 421 | |
| 9.34e-274 | 1 | 426 | 1 | 421 | |
| 1.09e-272 | 1 | 426 | 1 | 421 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.76e-62 | 37 | 420 | 32 | 389 | GPI mannosyltransferase 1 OS=Danio rerio OX=7955 GN=pigm PE=2 SV=1 |
|
| 5.48e-60 | 18 | 423 | 20 | 413 | GPI mannosyltransferase 1 OS=Gallus gallus OX=9031 GN=PIGM PE=2 SV=2 |
|
| 5.21e-57 | 5 | 422 | 9 | 416 | GPI mannosyltransferase 1 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GPI14 PE=3 SV=1 |
|
| 6.28e-56 | 6 | 412 | 25 | 427 | GPI mannosyltransferase 1 OS=Dictyostelium discoideum OX=44689 GN=pigm PE=3 SV=1 |
|
| 2.09e-54 | 37 | 423 | 41 | 417 | GPI mannosyltransferase 1 OS=Xenopus tropicalis OX=8364 GN=pigm PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000294 | 0.999681 | CS pos: 29-30. Pr: 0.8995 |
TMHMM Annotations download full data without filtering help
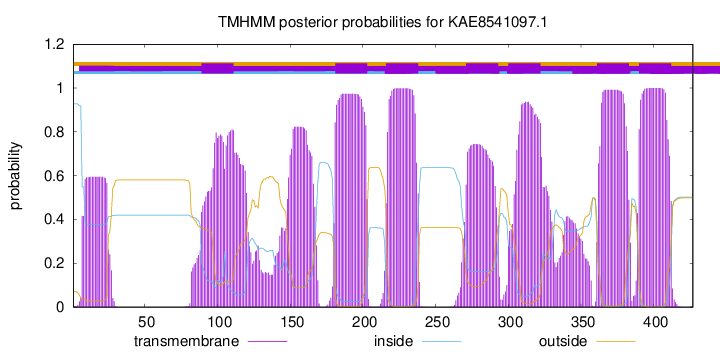
| Start | End |
|---|---|
| 89 | 111 |
| 181 | 203 |
| 216 | 238 |
| 271 | 293 |
| 300 | 322 |
| 361 | 383 |
| 390 | 412 |
