You are browsing environment: FUNGIDB
CAZyme Information: KAB8207508.1
You are here: Home > Sequence: KAB8207508.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8207508.1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | glycosyl hydrolase family 76-domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 474 | 1028 | 1.5e-148 | 0.9947183098591549 |
| GH76 | 24 | 391 | 4.1e-99 | 0.9664804469273743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 2.20e-179 | 29 | 372 | 2 | 347 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 225186 | BetA | 6.73e-87 | 468 | 1035 | 1 | 542 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 2.27e-85 | 472 | 1024 | 3 | 529 | choline dehydrogenase; Validated |
| 398739 | GMC_oxred_C | 2.40e-36 | 885 | 1022 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 3.01e-29 | 543 | 771 | 14 | 216 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1029 | 1 | 1021 | |
| 0.0 | 1 | 1029 | 1 | 1021 | |
| 0.0 | 468 | 1029 | 1 | 562 | |
| 0.0 | 517 | 1036 | 1 | 520 | |
| 1.44e-305 | 1 | 447 | 1 | 453 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.47e-85 | 28 | 381 | 35 | 404 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
|
| 3.77e-56 | 474 | 1026 | 1 | 561 | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid [Pleurotus eryngii] |
|
| 3.84e-56 | 474 | 1026 | 2 | 562 | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii [Pleurotus eryngii] |
|
| 4.26e-52 | 469 | 1030 | 1 | 573 | Crystal structure analysis of formate oxidase [Aspergillus oryzae RIB40],3Q9T_B Crystal structure analysis of formate oxidase [Aspergillus oryzae RIB40],3Q9T_C Crystal structure analysis of formate oxidase [Aspergillus oryzae RIB40] |
|
| 5.76e-52 | 469 | 1030 | 1 | 573 | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase [Aspergillus oryzae RIB40],5ZU3_B Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase [Aspergillus oryzae RIB40],5ZU3_C Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase [Aspergillus oryzae RIB40] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.74e-95 | 20 | 438 | 17 | 438 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
|
| 1.06e-90 | 23 | 393 | 18 | 401 | Putative mannan endo-1,6-alpha-mannosidase C970.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC970.02 PE=3 SV=1 |
|
| 2.31e-90 | 12 | 438 | 4 | 435 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
|
| 2.72e-87 | 29 | 437 | 29 | 440 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
|
| 3.91e-87 | 29 | 435 | 30 | 429 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
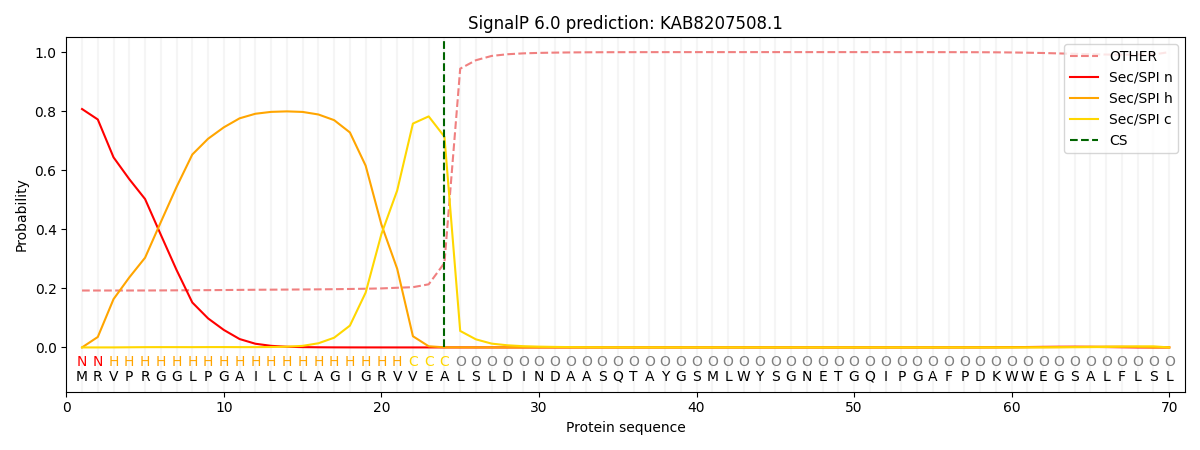
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.222747 | 0.777233 | CS pos: 24-25. Pr: 0.7136 |
