You are browsing environment: FUNGIDB
CAZyme Information: KAB8206568.1
You are here: Home > Sequence: KAB8206568.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
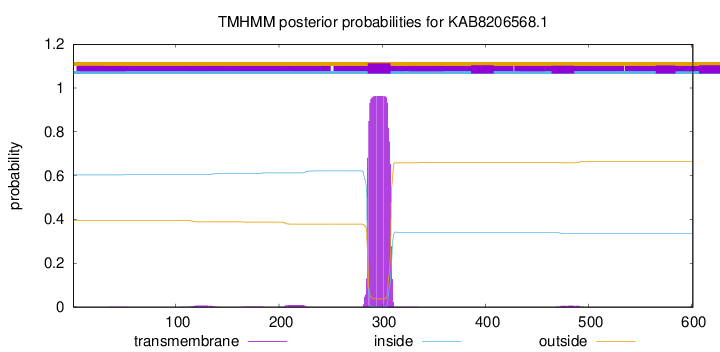
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8206568.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | N-acetylglucosaminylphosphatidylinositol deacetylase [Source:UniProtKB/TrEMBL;Acc:A0A5N6DN79] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396922 | PIG-L | 2.99e-21 | 328 | 449 | 1 | 125 | GlcNAc-PI de-N-acetylase. Members of this family are related to PIG-L an N-acetylglucosaminylphosphatidylinositol de-N-acetylase (EC:3.5.1.89) that catalyzes the second step in GPI biosynthesis. |
| 226297 | OCH1 | 4.30e-15 | 2 | 241 | 114 | 339 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 225031 | LmbE | 9.19e-13 | 320 | 482 | 7 | 172 | N-acetylglucosaminyl deacetylase, LmbE family [Carbohydrate transport and metabolism]. |
| 398274 | Gly_transf_sug | 1.88e-08 | 3 | 66 | 20 | 87 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 239211 | PAZ_piwi_like | 0.003 | 512 | 576 | 32 | 94 | PAZ domain, Piwi_like subfamily. In multi-cellular organisms, the Piwi protein appears to be essential for the maintenance of germline stem cells. In the Drosophila male germline, Piwi was shown to be involved in the silencing of retrotransposons in the male gametes. The Piwi proteins share their domain architecture with other members of the argonaute family. The PAZ domain has been named after the proteins Piwi, Argonaut, and Zwille. PAZ is found in two families of proteins that are essential components of RNA-mediated gene-silencing pathways, including RNA interference, the Piwi and Dicer families. PAZ functions as a nucleic acid binding domain, with a strong preference for single-stranded nucleic acids (RNA or DNA) or RNA duplexes with single-stranded 3' overhangs. It has been suggested that the PAZ domain provides a unique mode for the recognition of the two 3'-terminal nucleotides in single-stranded nucleic acids and buries the 3' OH group, and that it might recognize characteristic 3' overhangs in siRNAs within RISC (RNA-induced silencing) and other complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.87e-180 | 1 | 263 | 137 | 399 | |
| 4.15e-180 | 1 | 263 | 139 | 401 | |
| 6.15e-179 | 1 | 263 | 81 | 343 | |
| 6.29e-179 | 1 | 263 | 137 | 399 | |
| 6.29e-179 | 1 | 263 | 137 | 399 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.31e-29 | 324 | 484 | 32 | 191 | Probable N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gpi12 PE=3 SV=1 |
|
| 7.01e-28 | 21 | 249 | 197 | 394 | Initiation-specific alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=och1 PE=1 SV=2 |
|
| 2.52e-27 | 327 | 487 | 45 | 205 | N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase OS=Bos taurus OX=9913 GN=PIGL PE=2 SV=1 |
|
| 6.49e-25 | 327 | 479 | 44 | 197 | N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase OS=Homo sapiens OX=9606 GN=PIGL PE=1 SV=1 |
|
| 1.64e-24 | 327 | 479 | 44 | 197 | N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase OS=Rattus norvegicus OX=10116 GN=Pigl PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
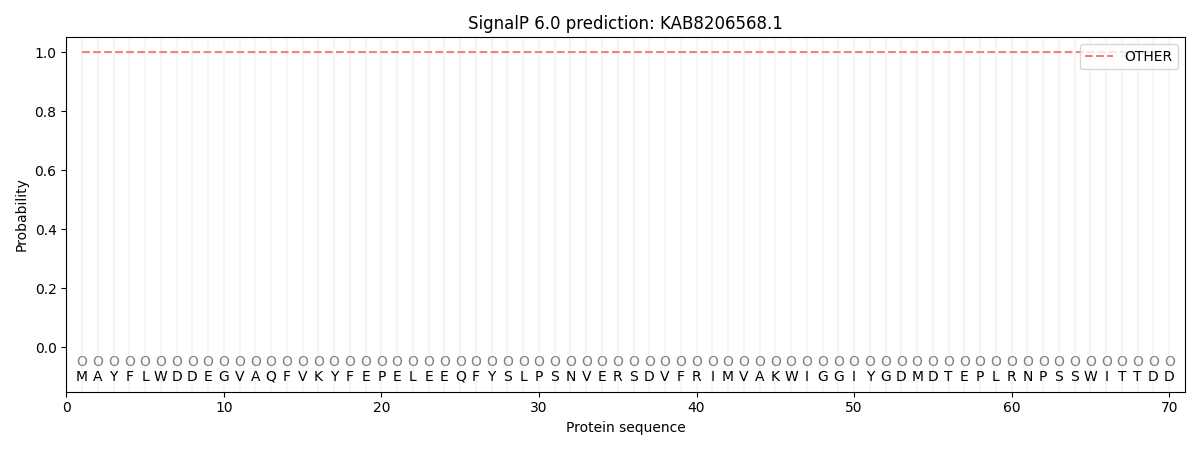
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000057 | 0.000000 |