You are browsing environment: FUNGIDB
CAZyme Information: KAB8205076.1
You are here: Home > Sequence: KAB8205076.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
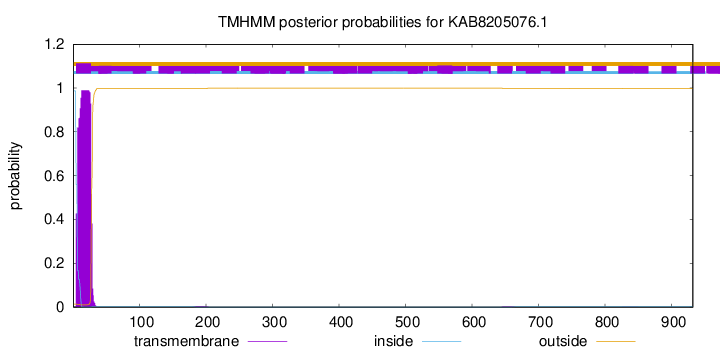
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8205076.1 | |||||||||||
| CAZy Family | PL26 | |||||||||||
| CAZyme Description | SET domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A5N6DIZ2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 264614; End:268997 Strand: - | |||||||||||
Full Sequence Download help
| MSRFICSGRM LGGTAILLST IIILIVFVNP PQTRILHSSS PQTASPTHNS SSPWNPSDVR | 60 |
| CSGLPDTSHI QVVVKTGTNI IYDKLPTQLL TALRCCQDPL IFADSEQDIG PYHVYDVLAN | 120 |
| VNETLKATHP NFAYYRTIKD YLSSGRDIRL LRTSRQAAWD LDKYKFIHML VETWERRPGH | 180 |
| DWYVFVEADT YLFWGNLVQW LARMDPAKPL YLGSAATFQN EKFAHGGSGV ILSREAMKRV | 240 |
| LDGDADLAAR YDERMHDEIY GDYVLMKALK EKGVELSNKW PMMQGEKQNT LPFGPGPNAG | 300 |
| SRHGCQPLIT MHSVTPVDVN TMWNYEQRRK HPQEPLLIGE LYDYFMGRAL PSQRDDWYNL | 360 |
| SDDLMFRAPG VEGQRQKSPA DMTPVEKEAY SSFEQFCRYH VLTCQGCVDL SNERVVPLSQ | 420 |
| LPPNVSETIC DKRDTTAAPE TNQDDHGPRP LKRRKTVSGL RLAPHNQDAA VTGTESQIDG | 480 |
| CAAPDSNGMT DDAQKSSVRQ VHSDTRFPQR KNPSKDTSAP VLEPTSTDKL IAGIWRQVFS | 540 |
| PVQLSRFHSV CLLELKRKVF RAVNTLCLKY YNQSQSSRAL EMIVQAYWIE CYEARIAVLR | 600 |
| LENPNLSAME IRMMGLREAC AVLNWKEKDL RNRIAIWRGY KEIKDAGGWA SLIFASAGVY | 660 |
| RFCKYRTGFG EGFSTRLRHI RSSLEVAADT LHPDWRDLLQ VIGQQETRQY HGHPHEWVTV | 720 |
| TGRPAVPLTS TYEHLQLPNG FHYRFIDECV LDTAAFGTED PRRVPEIDPG VCLVCKERQS | 780 |
| DEIEKNHCSC FPTLFGGVRN PVPVQLFHTT SGKNNGVIAR SNFDRGIAIG EFTGLITKGI | 840 |
| EGVDVMLGGT RTRTYQVFQG QMGNFTRFIN HSCRPNSQFQ RFYWRGKERI IVVSRGVTAG | 900 |
| SEITVDYSDH YWKQLNKICL CGEPCCRFRE RR | 932 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 380929 | SET_SETD2-like | 2.11e-15 | 860 | 927 | 68 | 136 | SET domain (including post-SET domain) found in SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2), ASH1-like protein (ASH1L) and similar proteins. This family includes SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2) and ASH1-like protein (ASH1L), which function as histone-lysine N-methyltransferases. SETD2 specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using demethylated 'Lys-36' (H3K36me2) as substrate. NSD2 shows histone H3 'Lys-27' (H3K27me) methyltransferase activity. ASH1L specifically methylates 'Lys-36' of histone H3 (H3K36me). The family also includes Arabidopsis thaliana ASH1-related protein 3 (ASHR3) and similar proteins. |
| 380949 | SET_SETD2 | 1.74e-14 | 863 | 927 | 73 | 138 | SET domain (including post-SET domain) found in SET domain-containing protein 2 (SETD2) and similar proteins. SETD2 (also termed HIF-1, huntingtin yeast partner B, huntingtin-interacting protein 1 (HIP-1), huntingtin-interacting protein B, lysine N-methyltransferase 3A or protein-lysine N-methyltransferase SETD2) acts as histone-lysine N-methyltransferase that specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using demethylated 'Lys-36' (H3K36me2) as substrate. It has been shown that methylation is a posttranslational modification of dynamic microtubules and that SETD2 methylates alpha-tubulin at lysine 40, the same lysine that is marked by acetylation on microtubules. Methylation of microtubules occurs during mitosis and cytokinesis and can be ablated by SETD2 deletion, which causes mitotic spindle and cytokinesis defects, micronuclei, and polyploidy. |
| 380922 | SET_Suv4-20-like | 6.01e-14 | 817 | 925 | 22 | 133 | SET domain (including post-SET domain) found in Drosophila melanogaster suppressor of variegation 4-20 (Suv4-20) and similar proteins. Suv4-20 (also termed Su(var)4-20) is a histone-lysine N-methyltransferase that specifically trimethylates 'Lys-20' of histone H4. It acts as a dominant suppressor of position-effect variegation. The family also includes Suv4-20 homologs, lysine N-methyltransferase 5B (KMT5B) and lysine N-methyltransferase 5C (KMT5C). Both KMT5B (also termed lysine-specific methyltransferase 5B, or suppressor of variegation 4-20 homolog 1, or Su(var)4-20 homolog 1, or Suv4-20h1) and KMT5C (also termed lysine-specific methyltransferase 5C, or suppressor of variegation 4-20 homolog 2, or Su(var)4-20 homolog 2, or Suv4-20h2) are histone methyltransferases that specifically trimethylate 'Lys-20' of histone H4 (H4K20me3). They play central roles in the establishment of constitutive heterochromatin in pericentric heterochromatin regions. |
| 214614 | SET | 3.31e-13 | 802 | 912 | 1 | 122 | SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain. Putative methyl transferase, based on outlier plant homologues |
| 380914 | SET | 4.31e-13 | 865 | 907 | 28 | 71 | SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain superfamily. The Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domain superfamily corresponds to SET domain-containing lysine methyltransferases, which catalyze site and state-specific methylation of lysine residues in histones that are fundamental in epigenetic regulation of gene activation and silencing in eukaryotic organisms. SET domains appear to be protein-protein interaction domains. It has been demonstrated that SET domains mediate interactions with a family of proteins that display similarity with dual-specificity phosphatases (dsPTPases). A subset of SET domains has been called PR domains. These domains are divergent in sequence from other SET domains, but also appear to mediate protein-protein interaction. The SET domain consists of two regions known as N-SET and C-SET. C-SET forms an unusual and conserved knot-like structure of probable functional importance. In addition to N-SET and C-SET, an insert region (I-SET) and flanking regions of high structural variability form part of the overall structure. Some family members contain a pre-SET domain, which is found in a number of histone methyltransferases (HMTase), and a post-SET domain, which harbors a zinc-binding site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW41467.1|GT31 | 1.60e-283 | 1 | 395 | 1 | 396 |
| QMW29395.1|GT31 | 1.60e-283 | 1 | 395 | 1 | 396 |
| QRD85744.1|GT31 | 1.60e-283 | 1 | 395 | 1 | 396 |
| BAE59144.1|GT31 | 1.60e-283 | 1 | 395 | 1 | 396 |
| UDD58664.1|GT31 | 1.51e-276 | 10 | 395 | 1 | 387 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YPU_A | 7.43e-09 | 861 | 927 | 146 | 214 | ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens],4YPU_B ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens] |
| 7BRE_B | 1.64e-08 | 816 | 927 | 38 | 161 | The crystal structure of MLL2 in complex with ASH2L and RBBP5 [Homo sapiens],7BRE_E The crystal structure of MLL2 in complex with ASH2L and RBBP5 [Homo sapiens] |
| 4YNP_A | 3.30e-08 | 861 | 927 | 146 | 214 | ASH1L SET domain S2259M mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens],4YNP_B ASH1L SET domain S2259M mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens] |
| 3OPE_A | 4.21e-08 | 861 | 927 | 142 | 210 | Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase [Homo sapiens],3OPE_B Structural Basis of Auto-inhibitory mechanism of Histone methyltransferase [Homo sapiens] |
| 4YPE_A | 4.45e-08 | 861 | 927 | 146 | 214 | ASH1L SET domain H2193F mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens],4YPE_B ASH1L SET domain H2193F mutant in complex with S-adenosyl methionine (SAM) [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P0CY36|Y1806_PHANO | 2.34e-129 | 470 | 930 | 119 | 601 | SET domain-containing protein SNOG_11806 OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=SNOG_11806 PE=3 SV=1 |
| sp|Q9Y7R4|SET1_SCHPO | 1.31e-06 | 863 | 927 | 852 | 916 | Histone-lysine N-methyltransferase, H3 lysine-4 specific OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=set1 PE=1 SV=1 |
| sp|Q18221|SET2_CAEEL | 1.47e-06 | 863 | 927 | 1439 | 1503 | Histone-lysine N-methyltransferase set-2 OS=Caenorhabditis elegans OX=6239 GN=set-2 PE=1 SV=2 |
| sp|Q9UMN6|KMT2B_HUMAN | 1.58e-06 | 816 | 927 | 2588 | 2711 | Histone-lysine N-methyltransferase 2B OS=Homo sapiens OX=9606 GN=KMT2B PE=1 SV=1 |
| sp|O08550|KMT2B_MOUSE | 1.58e-06 | 816 | 927 | 2586 | 2709 | Histone-lysine N-methyltransferase 2B OS=Mus musculus OX=10090 GN=Kmt2b PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
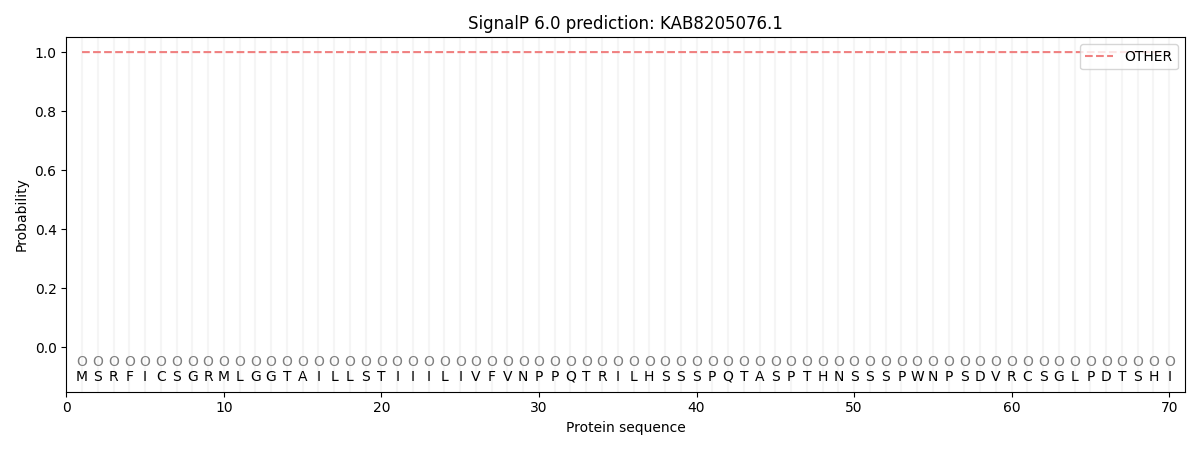
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999995 | 0.000032 |