You are browsing environment: FUNGIDB
CAZyme Information: KAB8204427.1
You are here: Home > Sequence: KAB8204427.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
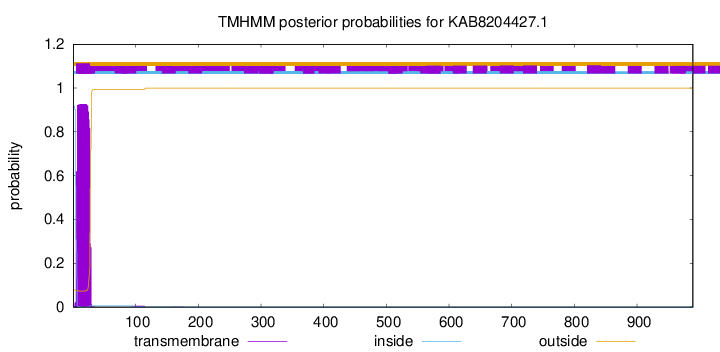
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8204427.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | GH115_C domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A5N6DHD7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.131:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH115 | 34 | 689 | 1.2e-236 | 0.8507890961262554 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 406396 | Glyco_hydro_115 | 1.15e-179 | 198 | 535 | 2 | 334 | Glycosyl hydrolase family 115. Glyco_hydro_115 is a family of glycoside hydrolases likely to have the activity of xylan a-1,2-glucuronidase, EC:3.2.1.131, or a-(4-O-methyl)-glucuronidase EC:3.2.1.-. |
| 407695 | GH115_C | 3.41e-61 | 808 | 985 | 1 | 172 | Gylcosyl hydrolase family 115 C-terminal domain. This domain is found at the C-terminus of glycosyl hydrolase family 115 proteins. This domain has a beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 989 | 1 | 989 | |
| 0.0 | 1 | 989 | 1 | 989 | |
| 0.0 | 1 | 989 | 1 | 989 | |
| 0.0 | 1 | 989 | 1 | 989 | |
| 0.0 | 1 | 989 | 1 | 999 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.41e-193 | 25 | 986 | 6 | 933 | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40],4ZMH_B Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T [Saccharophagus degradans 2-40] |
|
| 1.63e-128 | 18 | 708 | 28 | 708 | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C90_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_A Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus],4C91_B Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility [Bacteroides ovatus] |
|
| 7.46e-123 | 38 | 659 | 21 | 636 | Chain A, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
|
| 5.58e-121 | 38 | 659 | 20 | 635 | Chain A, xylan alpha-1,2-glucuronidase [uncultured bacterium] |
|
| 7.97e-116 | 55 | 989 | 32 | 966 | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112],6NPS_B Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus [Amphibacillus xylanus NBRC 15112] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000392 | 0.999586 | CS pos: 20-21. Pr: 0.9785 |