You are browsing environment: FUNGIDB
CAZyme Information: KAB8204241.1
You are here: Home > Sequence: KAB8204241.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8204241.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.14:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL20 | 98 | 285 | 1.3e-89 | 0.7991266375545851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404905 | Polysacc_lyase | 3.69e-68 | 39 | 272 | 2 | 213 | Polysaccharide lyase. This family includes heparin lyase I, EC:4.2.2.7. Heparin lyase I depolymerizes heparin by cleaving the glycosidic linkage next to an iduronic acid moiety. The structure of heparin lyase I consists of a beta-jelly roll domain with a long, deep substrate-binding groove and an unusual thumb domain containing many basic residues extending from the main body of the enzyme. This family also includes glucuronan lyase, EC:4.2.2.14. The structure glucuronan lyase is a beta-jelly roll. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.64e-202 | 10 | 285 | 20 | 295 | |
| 8.64e-202 | 10 | 285 | 20 | 295 | |
| 3.29e-200 | 16 | 285 | 1 | 270 | |
| 2.70e-199 | 16 | 285 | 1 | 270 | |
| 6.17e-196 | 10 | 285 | 405 | 680 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-86 | 34 | 284 | 1 | 237 | Chain A, Glucuronan lyase A [Trichoderma reesei] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
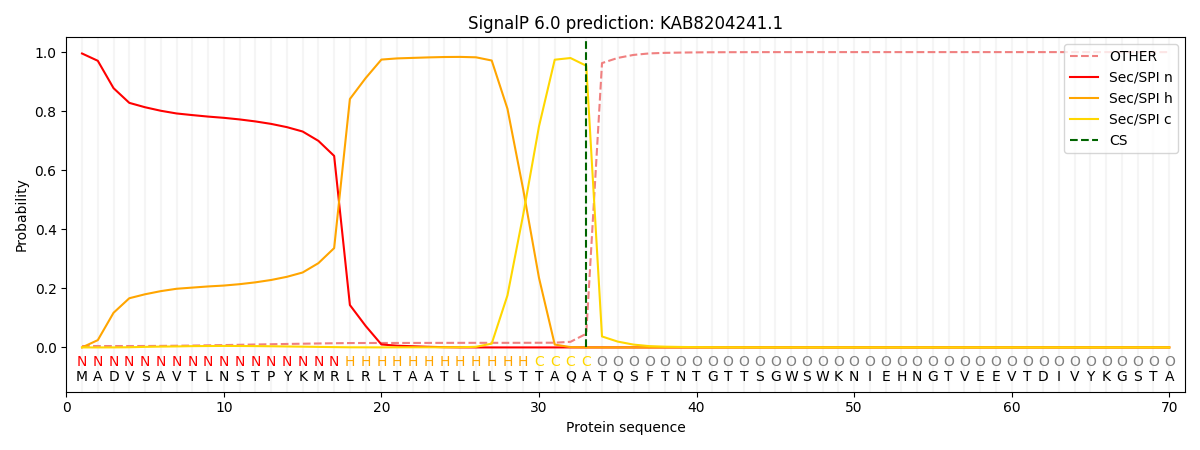
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.005308 | 0.994676 | CS pos: 33-34. Pr: 0.9537 |
