You are browsing environment: FUNGIDB
CAZyme Information: KAB8203930.1
Basic Information
help
| Species |
Aspergillus parasiticus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus
|
| CAZyme ID |
KAB8203930.1
|
| CAZy Family |
GH115 |
| CAZyme Description |
Peptidase_M14 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A5N6DIG1]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 973 |
ML734985|CGC1 |
107799.37 |
5.4475 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AparasiticusCBS117618 |
14005 |
N/A |
253 |
13752
|
|
| Gene Location |
No EC number prediction in KAB8203930.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
14 |
928 |
8.1e-68 |
0.9453883495145631 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 407266
|
Glyco_hydro_106 |
3.24e-14 |
240 |
604 |
399 |
755 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
11 |
967 |
29 |
992 |
| 7.57e-221 |
4 |
967 |
26 |
1013 |
| 1.06e-212 |
13 |
968 |
31 |
995 |
| 1.06e-212 |
13 |
968 |
31 |
995 |
| 1.49e-212 |
13 |
968 |
31 |
995 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.26e-07 |
179 |
584 |
407 |
799 |
Structure of Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
KAB8203930.1 has no Swissprot hit.
This protein is predicted as OTHER
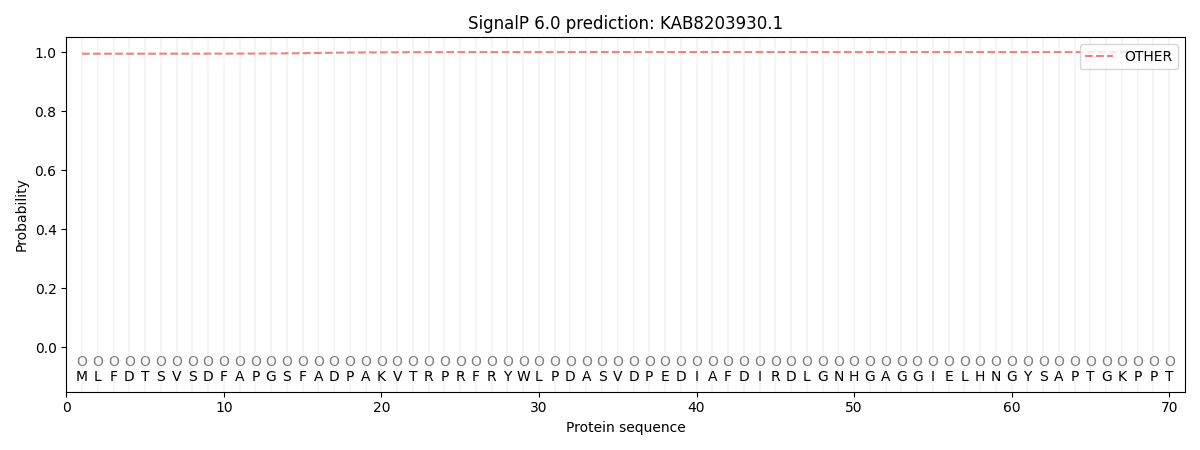
| Other |
SP_Sec_SPI |
CS Position |
| 0.995148 |
0.004897 |
|
There is no transmembrane helices in KAB8203930.1.

