You are browsing environment: FUNGIDB
CAZyme Information: KAB8203736.1
You are here: Home > Sequence: KAB8203736.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8203736.1 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | pectin lyase fold/virulence factor | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.171:11 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 67 | 210 | 2.4e-16 | 0.4553846153846154 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215540 | PLN03010 | 3.31e-10 | 41 | 210 | 48 | 222 | polygalacturonase |
| 403800 | Pectate_lyase_3 | 7.39e-09 | 41 | 205 | 3 | 151 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 215120 | PLN02188 | 5.94e-08 | 41 | 210 | 38 | 220 | polygalacturonase/glycoside hydrolase family protein |
| 177865 | PLN02218 | 1.32e-06 | 41 | 210 | 69 | 257 | polygalacturonase ADPG |
| 178580 | PLN03003 | 1.01e-05 | 41 | 210 | 25 | 203 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.15e-143 | 1 | 210 | 1 | 210 | |
| 4.15e-143 | 1 | 210 | 1 | 210 | |
| 4.15e-143 | 1 | 210 | 1 | 210 | |
| 4.15e-143 | 1 | 210 | 1 | 210 | |
| 3.38e-142 | 1 | 210 | 1 | 210 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.57e-84 | 20 | 210 | 1 | 190 | Rhamnogalacturonase A From Aspergillus Aculeatus [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.38e-144 | 1 | 210 | 1 | 210 | Probable rhamnogalacturonase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=rhgA PE=3 SV=1 |
|
| 6.00e-143 | 1 | 210 | 1 | 210 | Probable rhamnogalacturonase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=rhgA PE=3 SV=1 |
|
| 1.08e-106 | 9 | 210 | 7 | 208 | Rhamnogalacturonase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=rhgA PE=2 SV=2 |
|
| 3.64e-102 | 2 | 210 | 4 | 212 | Probable rhamnogalacturonase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=rhgB PE=3 SV=1 |
|
| 5.94e-101 | 1 | 210 | 3 | 212 | Rhamnogalacturonase B OS=Aspergillus niger OX=5061 GN=rhgB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
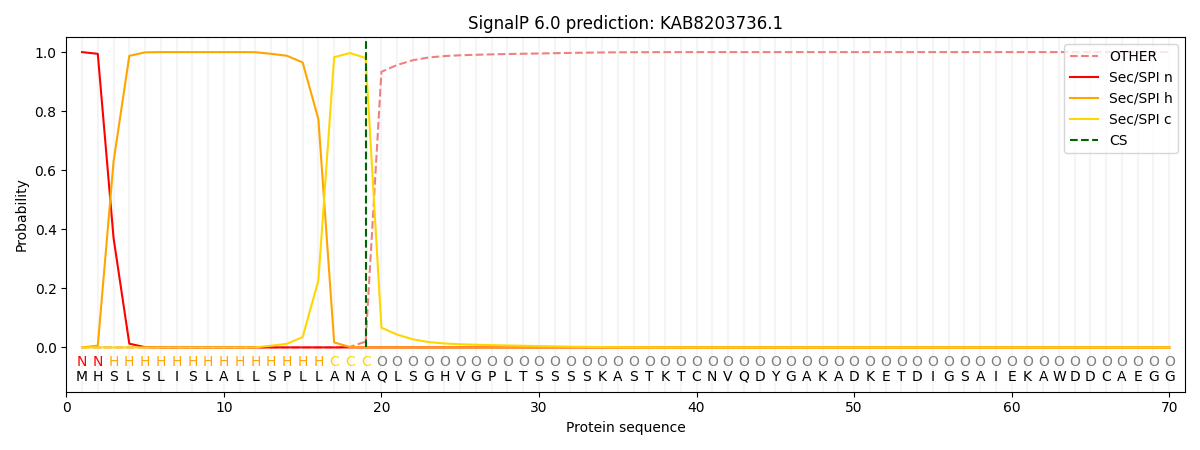
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000231 | 0.999755 | CS pos: 19-20. Pr: 0.9802 |
