You are browsing environment: FUNGIDB
CAZyme Information: KAB8200033.1
You are here: Home > Sequence: KAB8200033.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
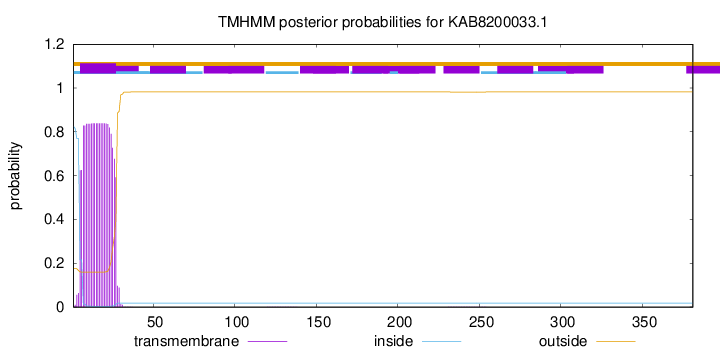
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8200033.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | pectin lyase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:25 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 115 | 299 | 5.5e-89 | 0.9893048128342246 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 2.04e-57 | 115 | 301 | 2 | 190 | Amb_all domain. |
| 366158 | Pec_lyase_C | 8.56e-16 | 125 | 297 | 30 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 226384 | PelB | 1.10e-14 | 28 | 381 | 35 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.57e-235 | 1 | 381 | 1 | 381 | |
| 7.57e-235 | 1 | 381 | 1 | 381 | |
| 7.57e-235 | 1 | 381 | 1 | 381 | |
| 7.57e-235 | 1 | 381 | 1 | 381 | |
| 7.57e-235 | 1 | 381 | 1 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.34e-182 | 26 | 380 | 6 | 359 | Pectin Lyase B [Aspergillus niger] |
|
| 1.08e-157 | 27 | 381 | 7 | 359 | Pectin Lyase A [Aspergillus niger] |
|
| 5.06e-156 | 27 | 381 | 7 | 359 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.34e-235 | 1 | 381 | 1 | 381 | Pectin lyase 1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pel1 PE=2 SV=1 |
|
| 3.50e-184 | 1 | 380 | 1 | 379 | Probable pectin lyase B OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pelB PE=3 SV=1 |
|
| 2.42e-182 | 1 | 381 | 1 | 380 | Probable pectin lyase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pelA PE=3 SV=1 |
|
| 2.61e-181 | 1 | 380 | 1 | 378 | Pectin lyase B OS=Aspergillus niger OX=5061 GN=pelB PE=1 SV=1 |
|
| 1.62e-170 | 1 | 381 | 1 | 378 | Probable pectin lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pelA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
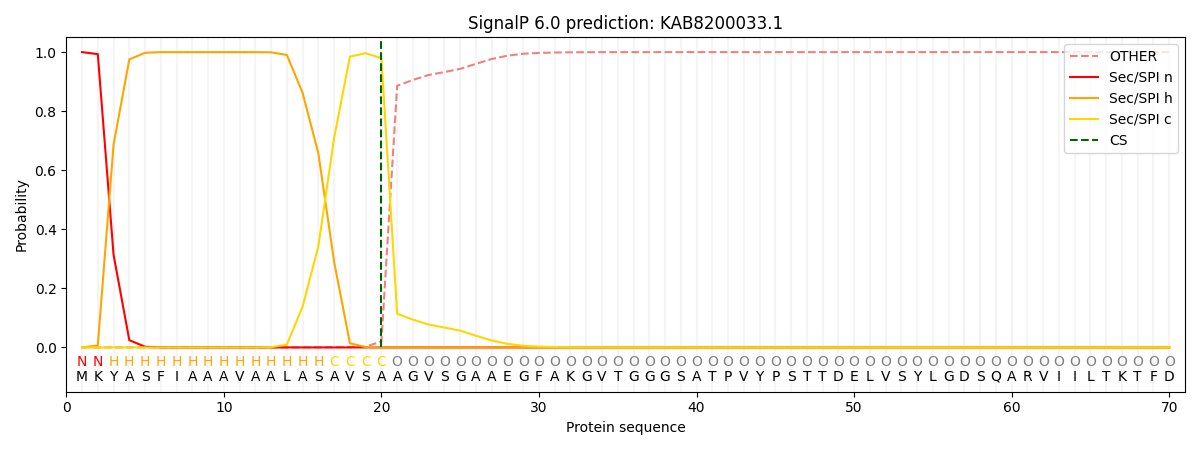
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000203 | 0.999774 | CS pos: 20-21. Pr: 0.9795 |