You are browsing environment: FUNGIDB
CAZyme Information: KAB8198925.1
You are here: Home > Sequence: KAB8198925.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus parasiticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus parasiticus | |||||||||||
| CAZyme ID | KAB8198925.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | glycoside hydrolase superfamily | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 25 | 302 | 3.5e-40 | 0.9935691318327974 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.11e-66 | 22 | 294 | 44 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 2.48e-40 | 24 | 302 | 1 | 309 | Glycosyl hydrolases family 17. |
| 409322 | T3SC_YbjN-like_2 | 0.004 | 98 | 143 | 89 | 133 | Uncharacterized protein is structurally similar to type III secretion system chaperones and YbjN family proteins. This family includes an uncharacterized protein from Methanothermobacter Thermautotrophicus that is structurally similar to type III secretion system (T3SS) chaperones (T3SC) that bind effector proteins, and is homologous to YbjN, a putative sensory transduction regulator protein found in Proteobacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.60e-226 | 1 | 304 | 1 | 304 | |
| 2.60e-226 | 1 | 304 | 1 | 304 | |
| 2.60e-226 | 1 | 304 | 1 | 304 | |
| 2.60e-226 | 1 | 304 | 1 | 304 | |
| 2.60e-226 | 1 | 304 | 1 | 304 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.03e-32 | 34 | 297 | 46 | 289 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 7.67e-32 | 34 | 297 | 46 | 289 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.03e-122 | 1 | 302 | 1 | 301 | Glucan 1,3-beta-glucosidase ARB_02797 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02797 PE=1 SV=1 |
|
| 1.19e-71 | 22 | 302 | 25 | 310 | Glucan 1,3-beta-glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=BGL2 PE=1 SV=1 |
|
| 1.66e-67 | 11 | 302 | 9 | 305 | Glucan 1,3-beta-glucosidase BGL2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BGL2 PE=1 SV=2 |
|
| 1.32e-66 | 11 | 302 | 9 | 305 | Glucan 1,3-beta-glucosidase OS=Candida albicans OX=5476 GN=BGL2 PE=3 SV=1 |
|
| 1.53e-62 | 15 | 302 | 35 | 318 | Glucan 1,3-beta-glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=bgl2 PE=2 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
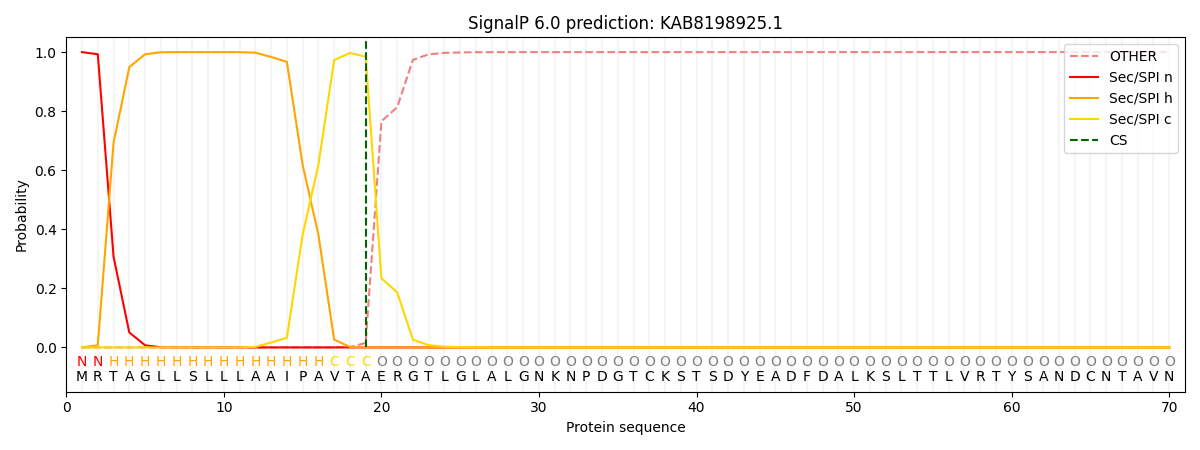
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000204 | 0.999745 | CS pos: 19-20. Pr: 0.9847 |
