You are browsing environment: FUNGIDB
CAZyme Information: KAA8909884.1
You are here: Home > Sequence: KAA8909884.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichomonascus ciferrii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Trichomonascaceae; Trichomonascus; Trichomonascus ciferrii | |||||||||||
| CAZyme ID | KAA8909884.1 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH132 | 37 | 309 | 6.8e-91 | 0.8415841584158416 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238033 | HECTc | 1.08e-125 | 1058 | 1422 | 2 | 352 | HECT domain; C-terminal catalytic domain of a subclass of Ubiquitin-protein ligase (E3). It binds specific ubiquitin-conjugating enzymes (E2), accepts ubiquitin from E2, transfers ubiquitin to substrate lysine side chains, and transfers additional ubiquitin molecules to the end of growing ubiquitin chains. |
| 397780 | SUN | 1.85e-124 | 39 | 267 | 1 | 230 | Beta-glucosidase (SUN family). Members of this family include Nca3, Sun4 and Sim1. This is a family of yeast proteins, involved in a diverse set of functions (DNA replication, aging, mitochondrial biogenesis and cell septation). BGLA from Candida wickerhamii has been characterized as a Beta-glucosidase EC:3.2.1.21. |
| 214523 | HECTc | 9.33e-116 | 1081 | 1421 | 4 | 328 | Domain Homologous to E6-AP Carboxyl Terminus with. E3 ubiquitin-protein ligases. Can bind to E2 enzymes. |
| 395507 | HECT | 2.39e-108 | 1109 | 1424 | 1 | 300 | HECT-domain (ubiquitin-transferase). The name HECT comes from Homologous to the E6-AP Carboxyl Terminus. |
| 227354 | HUL4 | 2.45e-102 | 1000 | 1424 | 452 | 872 | Ubiquitin-protein ligase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.36e-120 | 43 | 313 | 30 | 290 | |
| 5.23e-117 | 39 | 313 | 18 | 281 | |
| 5.23e-117 | 39 | 313 | 18 | 281 | |
| 7.26e-117 | 39 | 313 | 18 | 281 | |
| 1.40e-116 | 39 | 313 | 18 | 281 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-93 | 1021 | 1424 | 3 | 393 | Extended Hect domain of UBE3C E3 Ligase [Homo sapiens] |
|
| 1.44e-53 | 1053 | 1423 | 2 | 357 | STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY [Homo sapiens],1C4Z_B STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY [Homo sapiens],1C4Z_C STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY [Homo sapiens],1D5F_A STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY [Homo sapiens],1D5F_B STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY [Homo sapiens],1D5F_C STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY [Homo sapiens] |
|
| 1.26e-49 | 1053 | 1421 | 2132 | 2487 | Chain A, E3 ubiquitin-protein ligase HUWE1 [Nematocida sp. ERTm5] |
|
| 1.45e-48 | 1053 | 1421 | 2134 | 2489 | Chain A, E3 ubiquitin-protein ligase HUWE1 [Nematocida sp. ERTm5],7BII_B Chain B, E3 ubiquitin-protein ligase HUWE1 [Nematocida sp. ERTm5] |
|
| 1.73e-47 | 1011 | 1424 | 14 | 405 | Chain A, E3 ubiquitin-protein ligase HUWE1 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.34e-126 | 403 | 1424 | 9 | 1029 | Probable E3 ubiquitin protein ligase C167.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC167.07c PE=2 SV=1 |
|
| 7.20e-110 | 867 | 1424 | 350 | 910 | E3 ubiquitin-protein ligase HUL5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HUL5 PE=1 SV=1 |
|
| 8.09e-106 | 23 | 322 | 41 | 326 | Uncharacterized protein YMR244W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YMR244W PE=3 SV=1 |
|
| 1.44e-93 | 869 | 1424 | 621 | 1142 | E3 ubiquitin-protein ligase UPL7 OS=Arabidopsis thaliana OX=3702 GN=UPL7 PE=2 SV=1 |
|
| 2.38e-92 | 670 | 1424 | 314 | 1083 | Ubiquitin-protein ligase E3C OS=Mus musculus OX=10090 GN=Ube3c PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
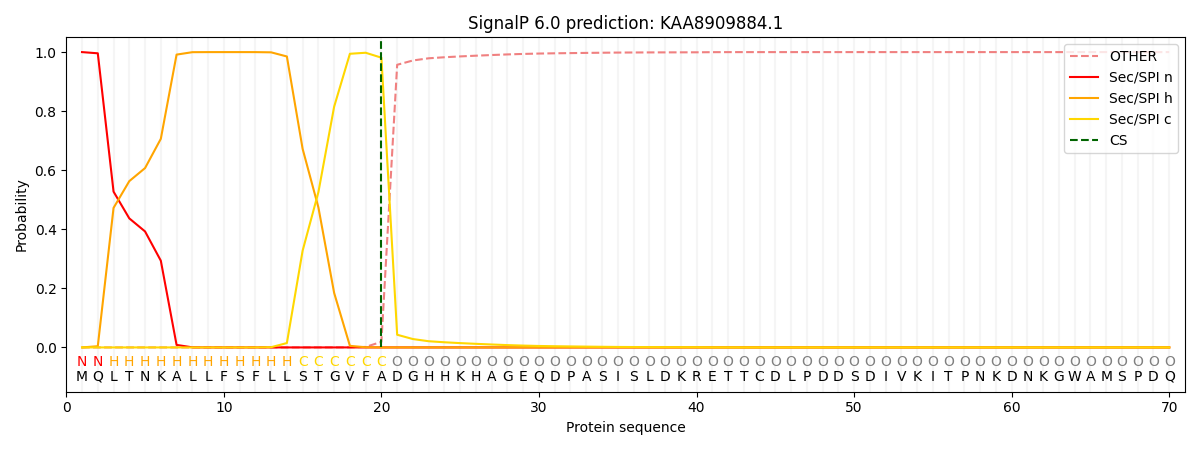
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000249 | 0.999748 | CS pos: 20-21. Pr: 0.9811 |
