You are browsing environment: FUNGIDB
CAZyme Information: KAA8904722.1
You are here: Home > Sequence: KAA8904722.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Trichomonascus ciferrii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Trichomonascaceae; Trichomonascus; Trichomonascus ciferrii | |||||||||||
| CAZyme ID | KAA8904722.1 | |||||||||||
| CAZy Family | GH132 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:26 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH72 | 237 | 483 | 3.7e-106 | 0.7788461538461539 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397351 | Glyco_hydro_72 | 2.35e-137 | 195 | 485 | 9 | 315 | Glucanosyltransferase. This is a family of glycosylphosphatidylinositol-anchored beta(1-3)glucanosyltransferases. The active site residues in the Aspergillus fumigatus example are the two glutamate residues at 160 and 261. |
| 404895 | DUF4267 | 1.04e-33 | 16 | 120 | 1 | 107 | Domain of unknown function (DUF4267). This family of integral membrane proteins is functionally uncharacterized. This family of proteins is found in bacteria and eukaryotes. Proteins in this family are typically between 126 and 142 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.92e-148 | 236 | 554 | 78 | 396 | |
| 8.05e-136 | 253 | 554 | 1 | 302 | |
| 2.13e-135 | 253 | 554 | 1 | 302 | |
| 1.13e-125 | 195 | 554 | 23 | 417 | |
| 3.07e-125 | 195 | 554 | 23 | 416 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.35e-64 | 205 | 481 | 79 | 340 | Saccharomyces cerevisiae Gas2p in complex with laminaripentaose [Saccharomyces cerevisiae],2W63_A Saccharomyces Cerevisiae Gas2p In Complex With Laminaritriose And Laminaritetraose [Saccharomyces cerevisiae],5O9O_A Crystal structure of ScGas2 in complex with compound 7. [Saccharomyces cerevisiae S288C],5O9P_A Crystal structure of Gas2 in complex with compound 10 [Saccharomyces cerevisiae S288C],5O9Q_A Crystal structure of ScGas2 in complex with compound 6 [Saccharomyces cerevisiae S288C],5O9R_A Crystal structure of ScGas2 in complex with compound 9 [Saccharomyces cerevisiae S288C],5O9Y_A Crystal structure of ScGas2 in complex with compound 11 [Saccharomyces cerevisiae S288C],5OA2_A Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA2_B Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA2_C Crystal structure of ScGas2 in complex with compound 8 [Saccharomyces cerevisiae S288C],5OA6_A Crystal structure of ScGas2 in complex with compound 12 [Saccharomyces cerevisiae S288C] |
|
| 3.62e-64 | 205 | 481 | 79 | 340 | Saccharomyces cerevisiae Gas2p apostructure (E176Q mutant) [Saccharomyces cerevisiae] |
|
| 3.62e-64 | 205 | 481 | 79 | 340 | SACCHAROMYCES CEREVISIAE GAS2P (E176Q MUTANT) IN COMPLEX WITH LAMINARITETRAOSE AND LAMINARIPENTAOSE [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-120 | 235 | 537 | 82 | 390 | 1,3-beta-glucanosyltransferase GAS4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS4 PE=1 SV=1 |
|
| 2.89e-86 | 188 | 560 | 15 | 415 | 1,3-beta-glucanosyltransferase gel2 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=gel2 PE=3 SV=1 |
|
| 2.89e-86 | 188 | 560 | 15 | 415 | 1,3-beta-glucanosyltransferase gel2 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=gel2 PE=1 SV=1 |
|
| 6.75e-76 | 235 | 532 | 83 | 382 | 1,3-beta-glucanosyltransferase GAS5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS5 PE=1 SV=1 |
|
| 8.08e-75 | 235 | 532 | 75 | 371 | 1,3-beta-glucanosyltransferase PGA4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PGA4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000035 | 0.000000 |
TMHMM Annotations download full data without filtering help
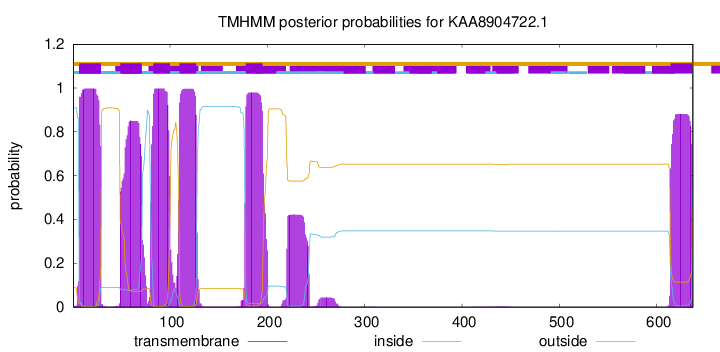
| Start | End |
|---|---|
| 7 | 29 |
| 49 | 71 |
| 83 | 100 |
| 110 | 127 |
| 178 | 195 |
| 614 | 636 |
