You are browsing environment: FUNGIDB
CAZyme Information: KAA8901156.1
You are here: Home > Sequence: KAA8901156.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Diutina rugosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; NA; Diutina; Diutina rugosa | |||||||||||
| CAZyme ID | KAA8901156.1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | Mannosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A642UN84] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 8 | 383 | 2.7e-68 | 0.987146529562982 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 1.01e-46 | 6 | 384 | 1 | 412 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 8.57e-09 | 5 | 362 | 39 | 426 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.68e-156 | 4 | 482 | 8 | 498 | |
| 4.24e-154 | 4 | 482 | 7 | 507 | |
| 1.33e-152 | 4 | 482 | 16 | 506 | |
| 5.06e-149 | 2 | 482 | 6 | 524 | |
| 2.88e-148 | 2 | 482 | 6 | 524 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.72e-156 | 4 | 482 | 8 | 498 | GPI mannosyltransferase 4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=SMP3 PE=3 SV=2 |
|
| 3.48e-133 | 4 | 482 | 9 | 535 | GPI mannosyltransferase 4 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=SMP3 PE=3 SV=2 |
|
| 5.28e-83 | 8 | 481 | 8 | 501 | GPI mannosyltransferase 4 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=SMP3 PE=3 SV=2 |
|
| 7.11e-79 | 4 | 468 | 2 | 490 | GPI mannosyltransferase 4 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=SMP3 PE=3 SV=2 |
|
| 5.84e-78 | 4 | 389 | 2 | 422 | GPI mannosyltransferase 4 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=smp3 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
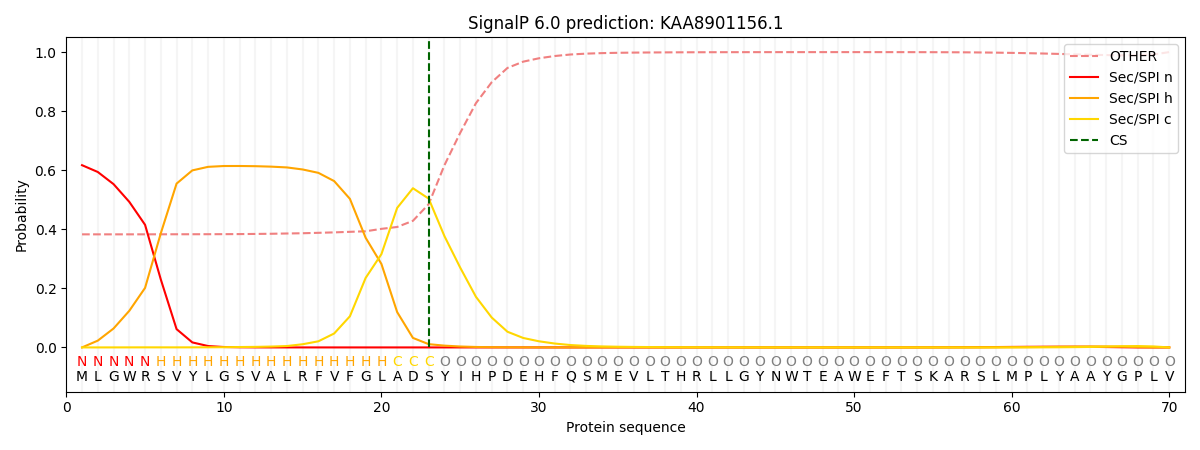
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.410115 | 0.589860 | CS pos: 23-24. Pr: 0.5038 |
TMHMM Annotations download full data without filtering help
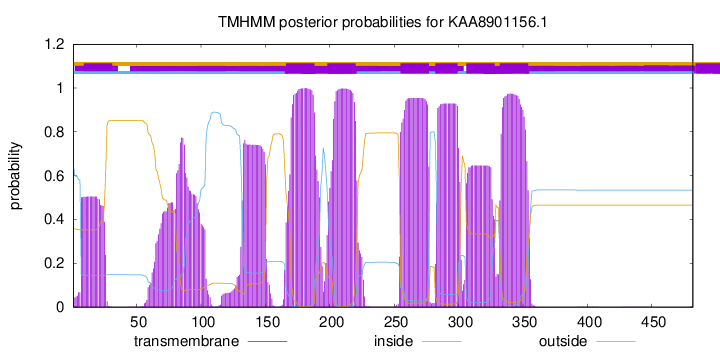
| Start | End |
|---|---|
| 166 | 188 |
| 198 | 220 |
| 255 | 277 |
| 282 | 299 |
| 306 | 328 |
| 332 | 354 |
