You are browsing environment: FUNGIDB
CAZyme Information: KAA8897150.1
You are here: Home > Sequence: KAA8897150.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Diutina rugosa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; NA; Diutina; Diutina rugosa | |||||||||||
| CAZyme ID | KAA8897150.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 8 | 390 | 4.8e-94 | 0.9922879177377892 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 6.59e-71 | 9 | 390 | 5 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 215437 | PLN02816 | 1.32e-67 | 5 | 493 | 40 | 541 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.89e-145 | 14 | 496 | 24 | 487 | |
| 1.03e-143 | 14 | 494 | 48 | 546 | |
| 5.24e-142 | 4 | 495 | 10 | 481 | |
| 1.17e-140 | 5 | 496 | 14 | 486 | |
| 1.75e-140 | 4 | 495 | 10 | 481 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.30e-143 | 4 | 495 | 10 | 481 | GPI mannosyltransferase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI10 PE=3 SV=2 |
|
| 9.61e-135 | 5 | 495 | 44 | 548 | GPI mannosyltransferase 3 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI10 PE=3 SV=2 |
|
| 8.89e-98 | 8 | 493 | 20 | 508 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
|
| 3.52e-88 | 11 | 491 | 57 | 526 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
|
| 5.69e-87 | 6 | 469 | 51 | 503 | GPI mannosyltransferase 3 OS=Xenopus laevis OX=8355 GN=pigb PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
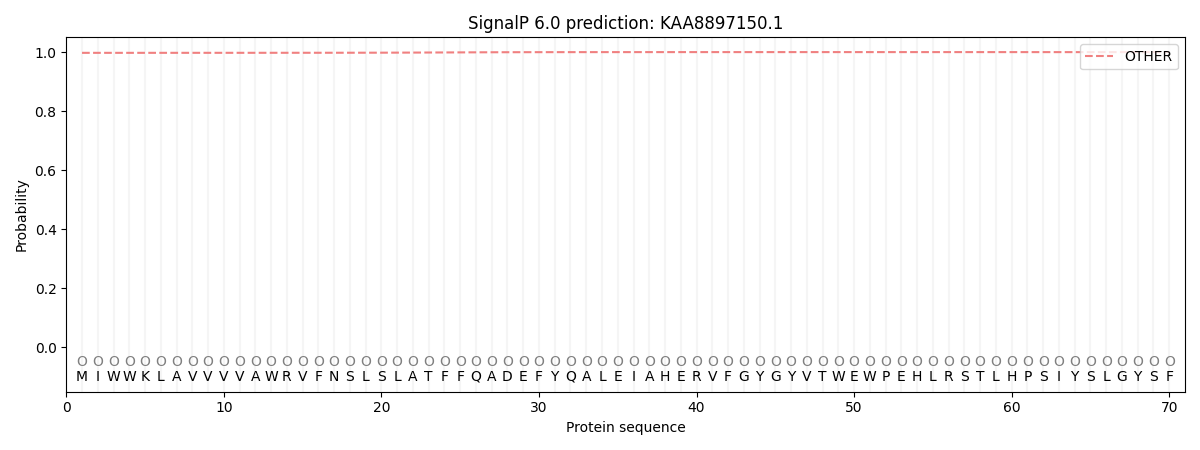
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.997609 | 0.002405 |
TMHMM Annotations download full data without filtering help
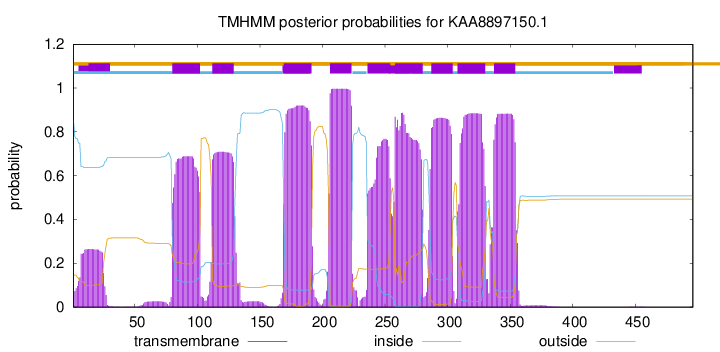
| Start | End |
|---|---|
| 80 | 102 |
| 112 | 129 |
| 169 | 191 |
| 206 | 223 |
| 236 | 254 |
| 258 | 280 |
| 287 | 304 |
| 308 | 330 |
| 337 | 354 |
