You are browsing environment: FUNGIDB
CAZyme Information: K441DRAFT_683579-t45_1-p1
You are here: Home > Sequence: K441DRAFT_683579-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
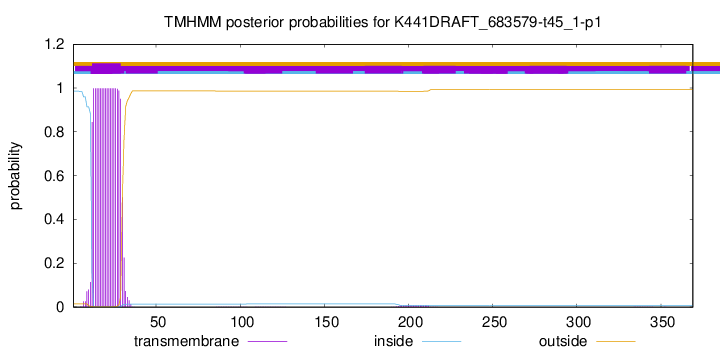
TMHMM annotations
Basic Information help
| Species | Cenococcum geophilum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Gloniaceae; Cenococcum; Cenococcum geophilum | |||||||||||
| CAZyme ID | K441DRAFT_683579-t45_1-p1 | |||||||||||
| CAZy Family | GH75 | |||||||||||
| CAZyme Description | glycosyltransferase family 62 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:2 | 2.4.1.257:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 72 | 334 | 6.6e-102 | 0.9776119402985075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 3.19e-152 | 71 | 335 | 1 | 265 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
| 224137 | GT2 | 5.76e-04 | 207 | 368 | 86 | 221 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.28e-190 | 3 | 369 | 2 | 367 | |
| 5.14e-187 | 4 | 369 | 3 | 367 | |
| 1.04e-186 | 3 | 369 | 2 | 367 | |
| 7.25e-185 | 4 | 369 | 7 | 378 | |
| 1.17e-168 | 2 | 369 | 3 | 374 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.14e-119 | 83 | 369 | 17 | 302 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.90e-118 | 83 | 369 | 107 | 392 | Mannan polymerase complexes subunit MNN9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN9 PE=1 SV=3 |
|
| 2.61e-117 | 83 | 369 | 84 | 368 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
|
| 4.58e-113 | 83 | 369 | 58 | 337 | Mannan polymerase complex subunit mnn9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnn9 PE=3 SV=1 |
|
| 6.97e-57 | 90 | 369 | 97 | 374 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 4.08e-48 | 90 | 369 | 190 | 514 | Mannan polymerase I complex VAN1 subunit OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=VAN1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
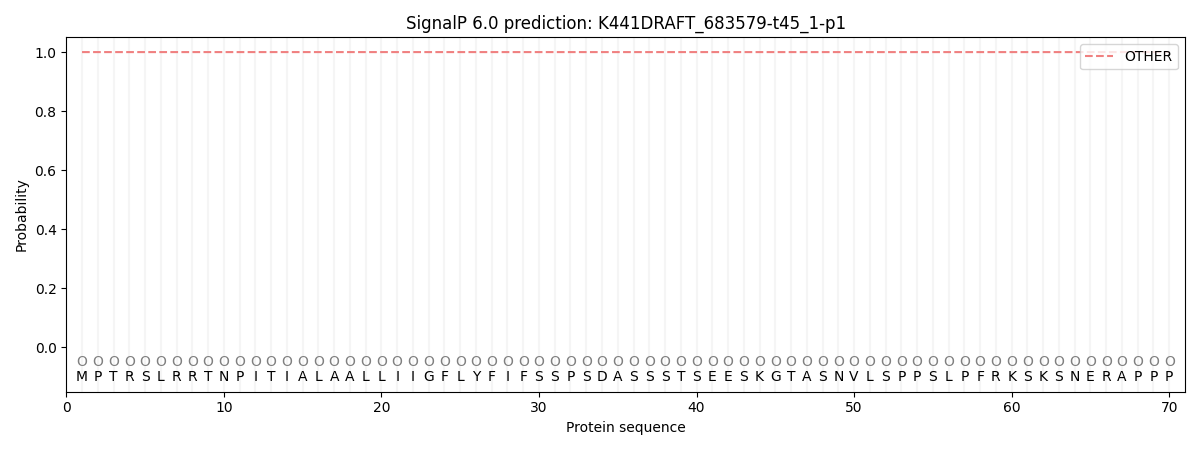
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000025 | 0.000006 |