You are browsing environment: FUNGIDB
CAZyme Information: K441DRAFT_671453-t45_1-p1
You are here: Home > Sequence: K441DRAFT_671453-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cenococcum geophilum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Gloniaceae; Cenococcum; Cenococcum geophilum | |||||||||||
| CAZyme ID | K441DRAFT_671453-t45_1-p1 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | glycoside hydrolase family 55 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 528 | 1280 | 2e-251 | 0.981081081081081 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403800 | Pectate_lyase_3 | 7.09e-74 | 559 | 786 | 1 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 403800 | Pectate_lyase_3 | 2.39e-08 | 915 | 982 | 1 | 74 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 227721 | Pgu1 | 3.77e-05 | 537 | 611 | 61 | 125 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| 215120 | PLN02188 | 0.001 | 561 | 840 | 38 | 311 | polygalacturonase/glycoside hydrolase family protein |
| 314993 | End_N_terminal | 0.005 | 567 | 595 | 1 | 28 | N terminal extension of bacteriophage endosialidase. This domain family is found in bacteria and viruses, and is approximately 70 amino acids in length. This domain is found in the bacteriophage protein endosialidase. The two N-terminal domains (this domain and the beta propeller) assemble in the compact 'cap' whereas the C-terminal domain forms an extended tail-like structure. The very N-terminal part of the 'cap' region (residues 246 to 312) holds the only alpha-helix of the protein and is presumably the residual part of the deleted N-terminal head-binding domain. The endosialidase protein complexes to form homotrimeric molecules. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1500 | 1 | 1476 | |
| 0.0 | 1 | 1500 | 1 | 1485 | |
| 0.0 | 343 | 1500 | 4 | 1153 | |
| 0.0 | 47 | 1471 | 47 | 1486 | |
| 0.0 | 22 | 1427 | 31 | 1497 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.10e-172 | 537 | 1270 | 26 | 738 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
|
| 1.88e-171 | 537 | 1279 | 6 | 744 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.49e-156 | 534 | 1260 | 40 | 854 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
|
| 1.47e-150 | 525 | 1267 | 36 | 764 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 3.30e-46 | 575 | 1250 | 71 | 719 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
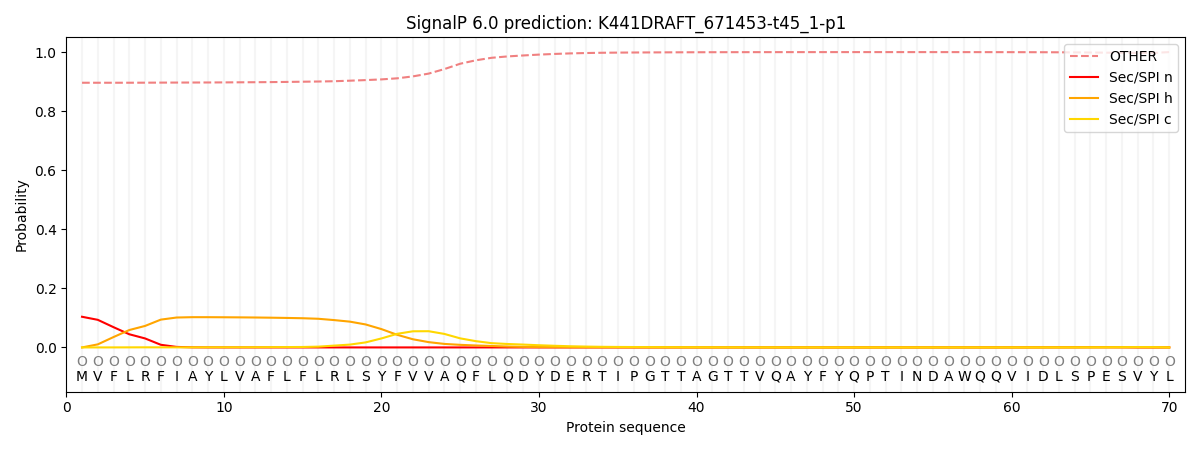
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.901531 | 0.098498 |

