You are browsing environment: FUNGIDB
CAZyme Information: K441DRAFT_661172-t45_1-p1
You are here: Home > Sequence: K441DRAFT_661172-t45_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
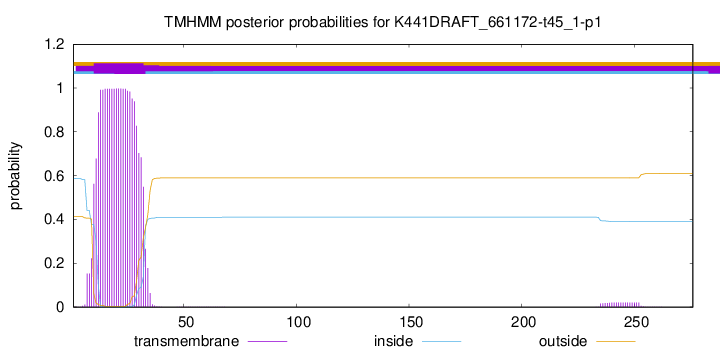
TMHMM annotations
Basic Information help
| Species | Cenococcum geophilum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Gloniaceae; Cenococcum; Cenococcum geophilum | |||||||||||
| CAZyme ID | K441DRAFT_661172-t45_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | glycosyltransferase family 34 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 49 | 250 | 4.8e-34 | 0.959349593495935 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 368535 | Glyco_transf_34 | 7.87e-26 | 59 | 249 | 22 | 233 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 215617 | PLN03182 | 7.22e-07 | 61 | 174 | 146 | 262 | xyloglucan 6-xylosyltransferase; Provisional |
| 215616 | PLN03181 | 4.92e-04 | 40 | 129 | 120 | 218 | glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.32e-101 | 1 | 272 | 1 | 272 | |
| 1.77e-96 | 39 | 271 | 65 | 297 | |
| 5.59e-89 | 49 | 272 | 52 | 275 | |
| 3.17e-83 | 49 | 272 | 55 | 275 | |
| 1.03e-45 | 30 | 265 | 63 | 307 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.17e-13 | 62 | 249 | 123 | 334 | Uncharacterized alpha-1,2-galactosyltransferase C1289.13c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1289.13c PE=3 SV=1 |
|
| 2.30e-13 | 67 | 249 | 112 | 315 | Alpha-1,2-galactosyltransferase gmh3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gmh3 PE=3 SV=1 |
|
| 1.76e-11 | 111 | 249 | 169 | 329 | Probable alpha-1,2-galactosyltransferase gmh2 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gmh2 PE=3 SV=1 |
|
| 3.79e-11 | 65 | 247 | 148 | 361 | Probable alpha-1,6-mannosyltransferase MNN10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN10 PE=1 SV=1 |
|
| 2.44e-10 | 68 | 270 | 110 | 329 | Probable alpha-1,2-galactosyltransferase gmh1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gmh1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.593045 | 0.406955 |