You are browsing environment: FUNGIDB
CAZyme Information: I317_06909-t43_1-p1
You are here: Home > Sequence: I317_06909-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
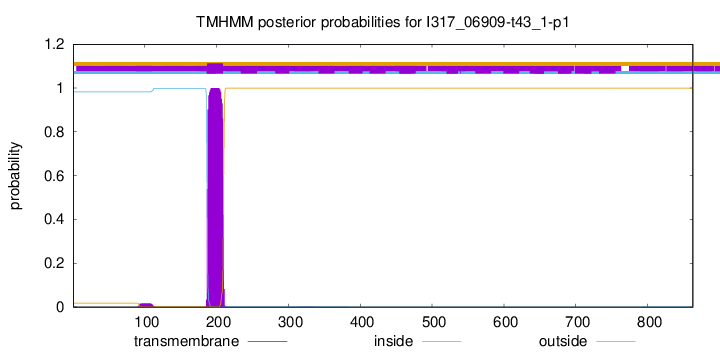
TMHMM annotations
Basic Information help
| Species | Kwoniella heveanensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella heveanensis | |||||||||||
| CAZyme ID | I317_06909-t43_1-p1 | |||||||||||
| CAZy Family | GT69 | |||||||||||
| CAZyme Description | glucan 1,3-beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 366 | 673 | 7.2e-100 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 1.29e-38 | 315 | 684 | 18 | 375 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 0.002 | 369 | 519 | 22 | 157 | Cellulase (glycosyl hydrolase family 5). |
| 130559 | ETRAMP | 0.002 | 181 | 217 | 45 | 82 | Plasmodium ring stage membrane protein ETRAMP. This model describes a family of proteins from the malaria parasite Plasmodium falciparum, several of which have been shown to be expressed specifically in the ring stage as well as the rident parasite Plasmodium yoelii. A homolog from Plasmodium chabaudi was localized to the parasitophorous vacuole membrane. Members have an initial hydrophobic, Phe/Tyr-rich stretch long enough to span the membrane, a highly charged region rich in Lys, a second putative transmembrane region, and a second highly charged, low complexity sequence region. Some members have up to 100 residues of additional C-terminal sequence. These genes have been shown to be found in the sub-telomeric regions of both P. falciparum and P. yoelii chromosomes |
| 411626 | FEA1_rel_lipo | 0.008 | 197 | 260 | 8 | 71 | FEA1-related lipoprotein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 26 | 839 | 12 | 819 | |
| 0.0 | 26 | 839 | 12 | 820 | |
| 0.0 | 26 | 839 | 12 | 820 | |
| 0.0 | 30 | 839 | 15 | 819 | |
| 0.0 | 26 | 839 | 12 | 815 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.25e-66 | 305 | 700 | 5 | 386 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 6.91e-66 | 307 | 700 | 1 | 380 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 7.92e-66 | 305 | 700 | 4 | 385 | F144Y/F258Y Double Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
|
| 1.13e-65 | 305 | 700 | 5 | 386 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
|
| 1.13e-65 | 305 | 700 | 5 | 386 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.33e-92 | 285 | 716 | 396 | 823 | Probable glucan 1,3-beta-glucosidase D OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgD PE=3 SV=1 |
|
| 2.33e-92 | 285 | 716 | 396 | 823 | Probable glucan 1,3-beta-glucosidase D OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=exgD PE=3 SV=2 |
|
| 6.34e-90 | 285 | 714 | 398 | 823 | Probable glucan 1,3-beta-glucosidase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgD PE=3 SV=1 |
|
| 6.34e-90 | 285 | 714 | 398 | 823 | Probable glucan 1,3-beta-glucosidase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=exgD PE=3 SV=1 |
|
| 6.35e-89 | 285 | 714 | 399 | 824 | Probable glucan 1,3-beta-glucosidase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
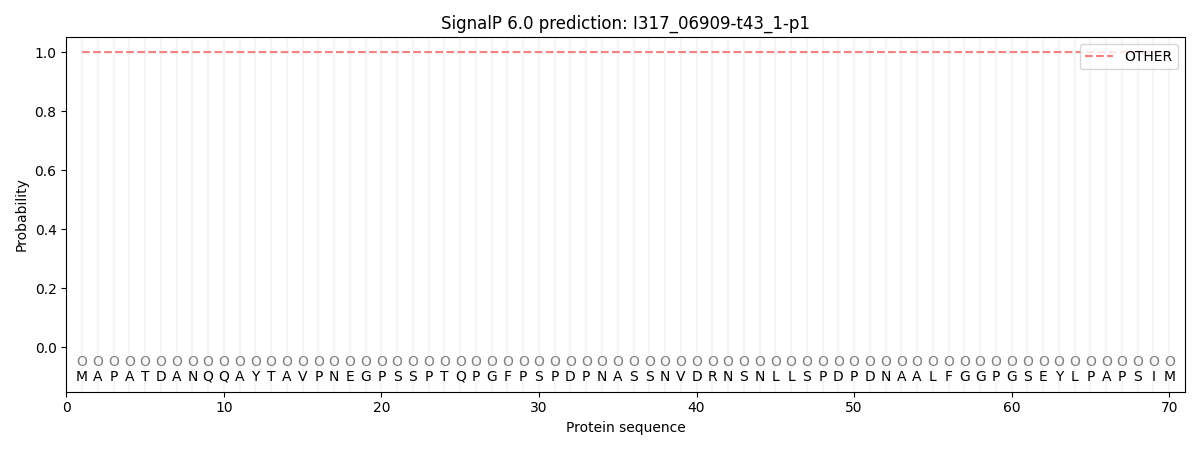
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000030 | 0.000003 |