You are browsing environment: FUNGIDB
CAZyme Information: I317_03095-t43_1-p1
You are here: Home > Sequence: I317_03095-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella heveanensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella heveanensis | |||||||||||
| CAZyme ID | I317_03095-t43_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | vesicle-fusing ATPase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA5 | 63 | 625 | 2.1e-228 | 0.9857904085257548 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 410912 | RecA-like_NSF-SEC18_r1-like | 1.21e-120 | 1007 | 1183 | 1 | 177 | first of two ATPase domains of NSF and SEC18, and similar ATPase domains. N-ethylmaleimide-sensitive factor (NSF) and Saccharomyces cerevisiae Vesicular-fusion protein Sec18, key factors for eukaryotic trafficking, are ATPases and SNARE disassembly chaperones. NSF/Sec18 activate or prime SNAREs, the terminal catalysts of membrane fusion. Sec18/NSF associates with SNARE complexes through binding Sec17/alpha-SNAP. Sec18 has an N-terminal cap domain and two nucleotide-binding domains (D1 and D2) which form the two rings of the hexameric complex. The hydrolysis of ATP by D1 generates most of the energy necessary to disassemble inactive SNARE bundles, while the D2 ring binds ATP to stabilize the homohexamer. This subfamily includes the first (D1) ATPase domain of NSF/Sec18, and belongs to the RecA-like NTPase family which includes the NTP binding domain of F1 and V1 H(+)ATPases, DnaB and related helicases as well as bacterial RecA and related eukaryotic and archaeal recombinases. The RecA-like NTPase family also includes bacterial conjugation proteins and related DNA transfer proteins involved in type II and type IV secretion. |
| 223540 | SpoVK | 1.18e-68 | 1030 | 1439 | 7 | 393 | AAA+-type ATPase, SpoVK/Ycf46/Vps4 family [Cell wall/membrane/envelope biogenesis, Cell cycle control, cell division, chromosome partitioning, Signal transduction mechanisms]. |
| 179699 | PRK03992 | 1.13e-64 | 999 | 1274 | 126 | 373 | proteasome-activating nucleotidase; Provisional |
| 273521 | CDC48 | 6.08e-64 | 999 | 1437 | 173 | 603 | AAA family ATPase, CDC48 subfamily. This subfamily of the AAA family ATPases includes two members each from three archaeal species. It also includes yeast CDC48 (cell division control protein 48) and the human ortholog, transitional endoplasmic reticulum ATPase (valosin-containing protein). These proteins in eukaryotes are involved in the budding and transfer of membrane from the transitional endoplasmic reticulum to the Golgi apparatus. |
| 223540 | SpoVK | 2.24e-63 | 999 | 1275 | 237 | 485 | AAA+-type ATPase, SpoVK/Ycf46/Vps4 family [Cell wall/membrane/envelope biogenesis, Cell cycle control, cell division, chromosome partitioning, Signal transduction mechanisms]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 61 | 627 | 37 | 605 | |
| 2.65e-315 | 14 | 636 | 5 | 614 | |
| 4.77e-313 | 61 | 627 | 37 | 605 | |
| 4.77e-313 | 61 | 627 | 37 | 605 | |
| 3.05e-225 | 64 | 628 | 41 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.13e-217 | 803 | 1526 | 5 | 734 | Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J94_B Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J94_C Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J94_D Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J94_E Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J94_F Structure of ATP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_A Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_B Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_C Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_D Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_E Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J95_F Structure of ADP-bound N-ethylmaleimide sensitive factor determined by single particle cryoelectron microscopy [Cricetulus griseus],3J96_A Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J96_B Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J96_C Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J96_D Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J96_E Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J96_F Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State I) [Cricetulus griseus],3J97_A Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J97_B Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J97_C Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J97_D Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J97_E Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J97_F Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State II) [Cricetulus griseus],3J98_A Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J98_B Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J98_C Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J98_D Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J98_E Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J98_F Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIa) [Cricetulus griseus],3J99_A Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus],3J99_B Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus],3J99_C Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus],3J99_D Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus],3J99_E Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus],3J99_F Structure of 20S supercomplex determined by single particle cryoelectron microscopy (State IIIb) [Cricetulus griseus] |
|
| 2.37e-216 | 803 | 1526 | 27 | 756 | NSF-D1D2 part in the whole 20S complex [Cricetulus griseus],6IP2_B NSF-D1D2 part in the whole 20S complex [Cricetulus griseus],6IP2_C NSF-D1D2 part in the whole 20S complex [Cricetulus griseus],6IP2_D NSF-D1D2 part in the whole 20S complex [Cricetulus griseus],6IP2_E NSF-D1D2 part in the whole 20S complex [Cricetulus griseus],6IP2_F NSF-D1D2 part in the whole 20S complex [Cricetulus griseus] |
|
| 2.48e-215 | 803 | 1526 | 26 | 755 | The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDM_B The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDM_C The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDM_D The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDM_E The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDM_F The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) [Cricetulus griseus],6MDN_A The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDN_B The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDN_C The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDN_D The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDN_E The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDN_F The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) [Cricetulus griseus],6MDO_A The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDO_B The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDO_C The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDO_D The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDO_E The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDO_F The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) [Cricetulus griseus],6MDP_A The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus],6MDP_B The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus],6MDP_C The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus],6MDP_D The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus],6MDP_E The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus],6MDP_F The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) [Cricetulus griseus] |
|
| 2.27e-66 | 1265 | 1526 | 6 | 258 | D2 Hexamerization Domain Of N-Ethylmaleimide Sensitive Factor (Nsf) [Cricetulus griseus] |
|
| 1.91e-65 | 1272 | 1526 | 15 | 259 | D2 Domain Of N-Ethylmaleimide-Sensitive Fusion Protein [Cricetulus griseus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-266 | 806 | 1529 | 59 | 789 | Vesicular-fusion protein sec18 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=sec18 PE=1 SV=1 |
|
| 7.17e-234 | 810 | 1527 | 31 | 756 | Vesicular-fusion protein SEC18 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SEC18 PE=1 SV=2 |
|
| 9.91e-220 | 813 | 1527 | 14 | 737 | Vesicle-fusing ATPase 2 OS=Drosophila melanogaster OX=7227 GN=Nsf2 PE=2 SV=2 |
|
| 1.36e-216 | 803 | 1526 | 2 | 731 | Vesicle-fusing ATPase OS=Homo sapiens OX=9606 GN=NSF PE=1 SV=3 |
|
| 1.92e-216 | 803 | 1526 | 2 | 731 | Vesicle-fusing ATPase OS=Pongo abelii OX=9601 GN=NSF PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
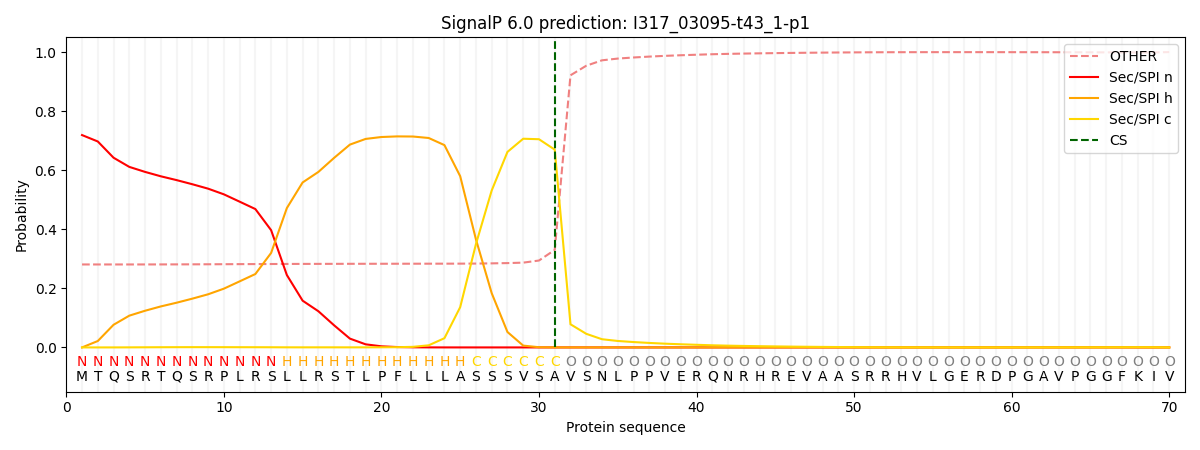
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.294036 | 0.705942 | CS pos: 31-32. Pr: 0.6697 |
