You are browsing environment: FUNGIDB
I311_00633-t41_1-p1
Basic Information
help
Species
Cryptococcus gattii VGI
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGI
CAZyme ID
I311_00633-t41_1-p1
CAZy Family
AA6
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
221
KN905585|CGC4 23585.57
7.0762
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CgattiiNT10
6987
1296108
243
6744
Gene Location
No EC number prediction in I311_00633-t41_1-p1.
Family
Start
End
Evalue
family coverage
PL14
1
174
5.8e-68
0.819047619047619
I311_00633-t41_1-p1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
6.27e-159
1
221
201
421
6.73e-148
1
221
176
396
4.66e-141
1
221
176
396
4.66e-141
1
221
176
396
2.24e-140
1
221
181
401
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
7.53e-41
1
174
93
270
Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai]
more
I311_00633-t41_1-p1 has no Swissprot hit.
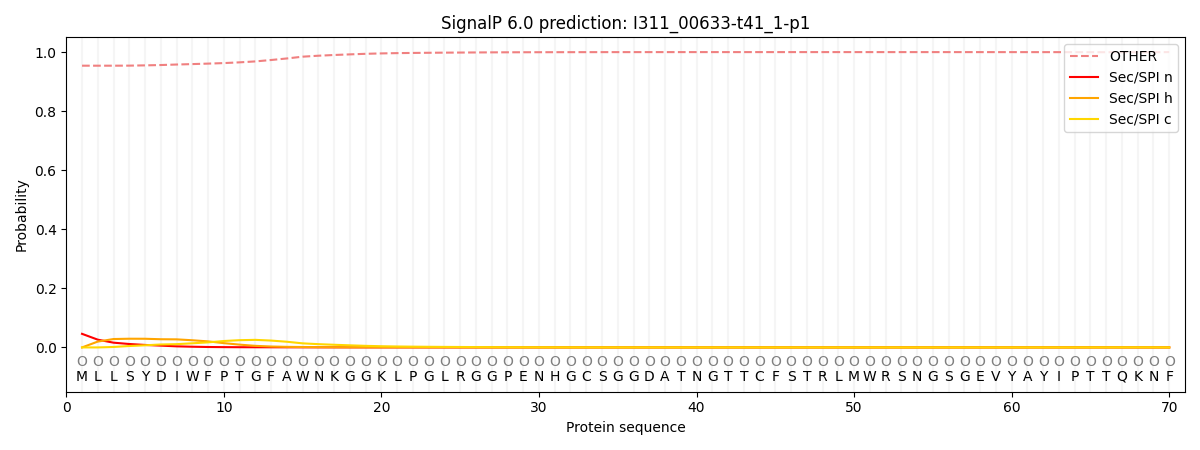
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.956989
0.043023
There is no transmembrane helices in I311_00633-t41_1-p1.