You are browsing environment: FUNGIDB
I308_00910-t35_1-p1
Basic Information
help
Species
Cryptococcus gattii VGIV
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGIV
CAZyme ID
I308_00910-t35_1-p1
CAZy Family
CBM67|GH78
CAZyme Description
alginate lyase
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
309
KN848856|CGC9 32362.06
8.4086
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CgattiiIND107
6843
1296105
147
6696
Gene Location
No EC number prediction in I308_00910-t35_1-p1.
Family
Start
End
Evalue
family coverage
PL14
51
262
1.1e-74
0.9857142857142858
I308_00910-t35_1-p1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
5.84e-195
2
309
114
421
7.54e-192
34
309
121
396
1.28e-184
34
309
126
401
7.18e-183
34
309
121
396
7.18e-183
34
309
121
396
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.71e-45
53
262
59
270
Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai]
more
3.47e-11
76
262
61
237
Structure of alginate lyase Aly36B [Chitinophaga sp. MD30]
more
I308_00910-t35_1-p1 has no Swissprot hit.
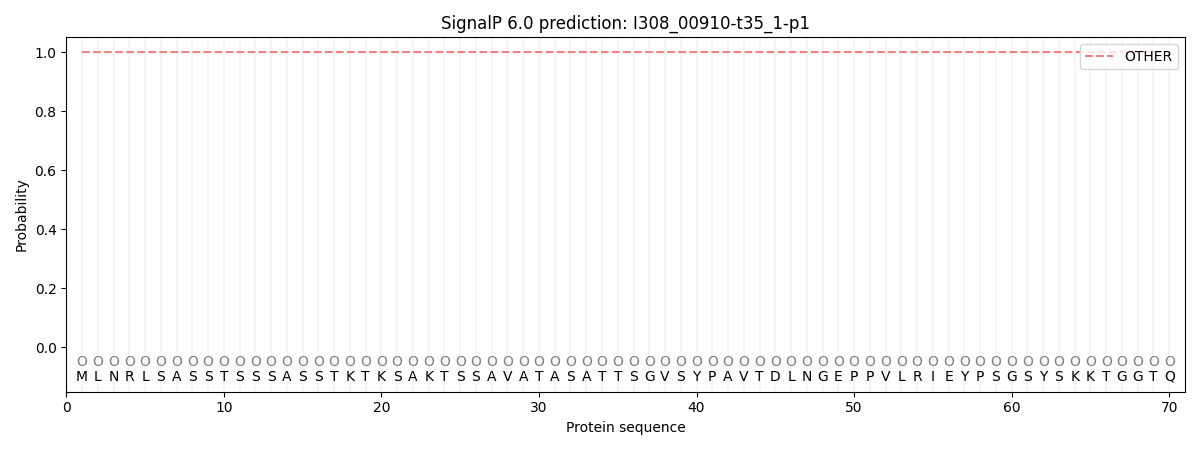
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000042
0.000006
There is no transmembrane helices in I308_00910-t35_1-p1.