You are browsing environment: FUNGIDB
CAZyme Information: I306_00441-t38_1-p1
You are here: Home > Sequence: I306_00441-t38_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
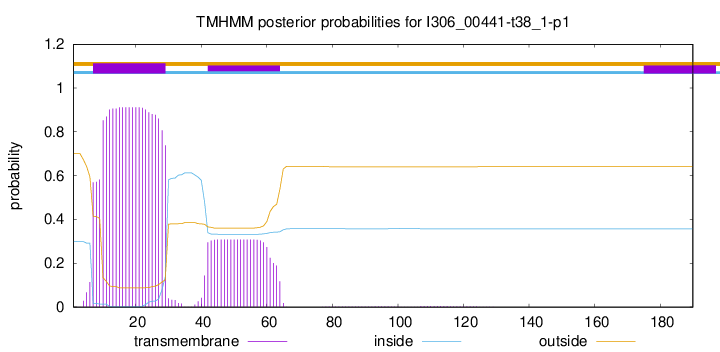
TMHMM annotations
Basic Information help
| Species | Cryptococcus gattii VGI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGI | |||||||||||
| CAZyme ID | I306_00441-t38_1-p1 | |||||||||||
| CAZy Family | AA5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH152 | 36 | 186 | 2.2e-29 | 0.7222222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395248 | Thaumatin | 8.12e-27 | 36 | 183 | 1 | 149 | Thaumatin family. |
| 185758 | TLP-F | 3.10e-20 | 32 | 183 | 1 | 158 | thaumatin-like proteins: basidiomycete homologs. This subfamily is represented by Lentinula edodes TLG1, a thaumatin-like protein (TLP), as well as, other basidiomycete homologs. In general, TLPs are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. TLG1 TLP is involved in lentinan degradation and fruiting body senescence. TLG1 expressed in Escherichia coli and Aspergillus oryzae exhibited beta-1,3-glucanase activity and demonstrated lentinan degrading activity. TLG1 is proposed to be involved in lentinan and cell wall degradation during senescence following harvest and spore diffusion. TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. TLG1 from Lentinula edodes contains the required acidic amino acids conserved in the appropriate positions to possess an electronegative cleft. TLPs within this subfamily contain 13 conserved Cys residues; the number of total Cys residues in these TLPs varies from 16 in L. edodes TLG1 to 18 in other basidiomycete homologs. |
| 185757 | TLP-PA | 3.36e-19 | 31 | 183 | 1 | 158 | allergenic/antifungal thaumatin-like proteins: plant and animal homologs. This subfamily is represented by the thaumatin-like proteins (TLPs), Cherry Allergen Pru Av 2 TLP, Peach PpAZ44 TLP (a propylene-induced TLP in abscission), the Caenorhabditis elegans thaumatin family member (thn-6), and other plant and animal homologs. TLPs are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Due to their inducible expression by environmental stresses such as pathogen/pest attack, drought and cold, plant TLPs are classified as the pathogenesis-related (PR) protein family 5 (PR5). Several members of the plant TLP family have been reported as food allergens from fruits (i.e., cherry, Pru av 2; bell pepper, Cap a1; tomatoes, Lyc e NP24) and pollen allergens from conifers (i.e., mountain cedar, Jun a 3; Arizona cypress, Cup a3; Japanese cedar, Cry j3). TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. It has been proposed that the antifungal activity of plant PR5 proteins relies on the strong electronegative character of this cleft. Some TLPs hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. TLPs within this subfamily contain 16 conserved Cys residues. |
| 128501 | THN | 2.16e-17 | 32 | 183 | 1 | 156 | Thaumatin family. The thaumatin family gathers proteins related to plant pathogenesis. The thaumatin family includes very basic members with extracellular and vacuolar localization. Thaumatin itsel is a potent sweet-tasting protein. Several members of this family display significant in vitro activity of inhibiting hyphal growth or spore germination of various fungi probably by a membrane permeabilizing mechanism. |
| 185756 | TLP-P | 1.62e-16 | 32 | 181 | 1 | 141 | thaumatin and allergenic/antifungal thaumatin-like proteins: plant homologs. This subfamily is represented by the sweet-tasting protein thaumatin from the African berry Thaumatococcus daniellii, allergenic/antifungal Thaumatin-like proteins (TLPs), and related plant proteins. TLPs are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Plant TLPs are classified as pathogenesis-related (PR) protein family 5 (PR5), their expression is induced by environmental stresses such as pathogen/pest attack, drought and cold. TLPs in this subfamily include such proteins as zeamatin, found in high concentrations in cereal seeds, and osmotin, a salt-induced protein in osmotically stressed plants. Several members of the plant TLP family have been reported as food allergens from fruits (i.e., cherry, Pru av 2; bell pepper, Cap a1; tomatoes, Lyc e NP24) and pollen allergens from conifers (i.e., mountain cedar, Jun a 3; Arizona cypress, Cup a3; Japanese cedar, Cry j3). Thaumatin and TLPs are three-domain, crescent-fold structures with either an electronegative, electropositive, or neutral cleft occurring between domains I and II. It has been proposed that the antifungal activity of plant PR5 proteins relies on the strong electronegative character of this cleft. IgE-binding epitopes of mountain Cedar (Juniperus ashei) allergen Jun a 3, which interact with pooled IgE from patients suffering allergenic response to this allergen, were mainly located on the helical domain II; the best-conserved IgE-binding epitope predicted for TLPs corresponds to this region. Some TLPs hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. Most TLPs contain 16 conserved Cys residues. A deletion within the third domain (domain II) of the Triticum aestivum thaumatin-like xylanase inhibitor is observed, thus, only 10 conserved Cys residues are present within this smaller TLP and similar homologs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.44e-113 | 1 | 188 | 1 | 198 | |
| 4.17e-104 | 1 | 188 | 1 | 198 | |
| 4.01e-103 | 1 | 188 | 1 | 198 | |
| 4.01e-103 | 1 | 188 | 1 | 198 | |
| 8.60e-84 | 1 | 188 | 1 | 181 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.28e-11 | 75 | 183 | 48 | 160 | High resolution structure of a cherry allergen Pru av 2 [Prunus avium] |
|
| 9.03e-11 | 34 | 183 | 5 | 143 | Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7A [Musa acuminata] |
|
| 1.37e-10 | 34 | 183 | 5 | 149 | Structure of HYPER-VIL-thaumatin [Thaumatococcus daniellii] |
|
| 1.89e-10 | 34 | 183 | 5 | 149 | Structure K137A thaumatin [Thaumatococcus daniellii] |
|
| 1.89e-10 | 34 | 183 | 5 | 149 | Structure of hyper-sweet thaumatin (D21N) [Thaumatococcus daniellii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-15 | 16 | 183 | 14 | 195 | Thaumatin-like protein 1 OS=Arabidopsis thaliana OX=3702 GN=TLP1 PE=2 SV=1 |
|
| 7.64e-14 | 25 | 183 | 18 | 170 | Pathogenesis-related thaumatin-like protein 3.3 (Fragment) OS=Cryptomeria japonica OX=3369 PE=1 SV=1 |
|
| 1.65e-13 | 33 | 180 | 30 | 166 | Thaumatin-like protein OS=Oryza sativa subsp. japonica OX=39947 GN=Os12g0628600 PE=1 SV=1 |
|
| 3.85e-13 | 11 | 181 | 4 | 159 | Thaumatin-like pathogenesis-related protein 4 OS=Avena sativa OX=4498 GN=RASTL-4 PE=2 SV=1 |
|
| 9.10e-13 | 7 | 167 | 4 | 160 | Pathogenesis-related thaumatin-like protein 3.6 (Fragment) OS=Cryptomeria japonica OX=3369 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
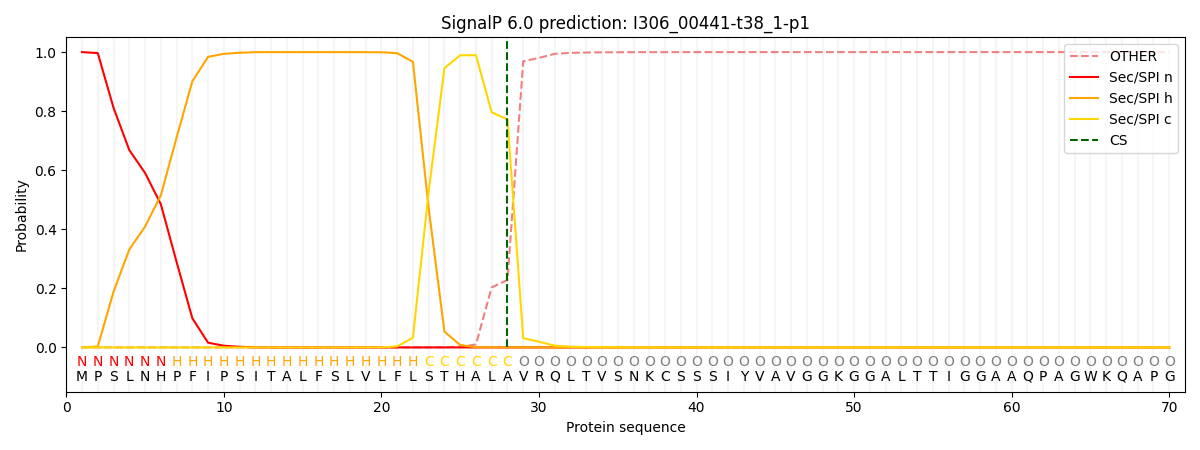
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000498 | 0.999475 | CS pos: 28-29. Pr: 0.7718 |