You are browsing environment: FUNGIDB
CAZyme Information: I303_06171-t42_1-p1
You are here: Home > Sequence: I303_06171-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella dejecticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella dejecticola | |||||||||||
| CAZyme ID | I303_06171-t42_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT47 | 796 | 1086 | 1.6e-39 | 0.9831081081081081 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397245 | Exostosin | 7.58e-54 | 793 | 1086 | 2 | 290 | Exostosin family. The EXT family is a family of tumor suppressor genes. Mutations of EXT1 on 8q24.1, EXT2 on 11p11-13, and EXT3 on 19p have been associated with the autosomal dominant disorder known as hereditary multiple exostoses (HME). This is the most common known skeletal dysplasia. The chromosomal locations of other EXT genes suggest association with other forms of neoplasia. EXT1 and EXT2 have both been shown to encode a heparan sulphate polymerase with both D-glucuronyl (GlcA) and N-acetyl-D-glucosaminoglycan (GlcNAC) transferase activities. The nature of the defect in heparan sulphate biosynthesis in HME is unclear. |
| 223528 | WcaG | 2.47e-33 | 90 | 412 | 1 | 306 | Nucleoside-diphosphate-sugar epimerase [Cell wall/membrane/envelope biogenesis]. |
| 396097 | Epimerase | 5.18e-22 | 93 | 331 | 2 | 231 | NAD dependent epimerase/dehydratase family. This family of proteins utilize NAD as a cofactor. The proteins in this family use nucleotide-sugar substrates for a variety of chemical reactions. |
| 212494 | SDR_e | 5.40e-22 | 93 | 347 | 2 | 199 | extended (e) SDRs. Extended SDRs are distinct from classical SDRs. In addition to the Rossmann fold (alpha/beta folding pattern with a central beta-sheet) core region typical of all SDRs, extended SDRs have a less conserved C-terminal extension of approximately 100 amino acids. Extended SDRs are a diverse collection of proteins, and include isomerases, epimerases, oxidoreductases, and lyases; they typically have a TGXXGXXG cofactor binding motif. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold, an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Sequence identity between different SDR enzymes is typically in the 15-30% range; they catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase numbering). In addition to the Tyr and Lys, there is often an upstream Ser and/or an Asn, contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Atypical SDRs generally lack the catalytic residues characteristic of the SDRs, and their glycine-rich NAD(P)-binding motif is often different from the forms normally seen in classical or extended SDRs. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. |
| 187551 | UDP_G4E_3_SDR_e | 1.16e-16 | 93 | 403 | 2 | 295 | UDP-glucose 4 epimerase (G4E), subgroup 3, extended (e) SDRs. Members of this bacterial subgroup are identified as possible sugar epimerases, such as UDP-glucose 4 epimerase. However, while the NAD(P)-binding motif is fairly well conserved, not all members retain the canonical active site tetrad of the extended SDRs. UDP-glucose 4 epimerase (aka UDP-galactose-4-epimerase), is a homodimeric extended SDR. It catalyzes the NAD-dependent conversion of UDP-galactose to UDP-glucose, the final step in Leloir galactose synthesis. Extended SDRs are distinct from classical SDRs. In addition to the Rossmann fold (alpha/beta folding pattern with a central beta-sheet) core region typical of all SDRs, extended SDRs have a less conserved C-terminal extension of approximately 100 amino acids. Extended SDRs are a diverse collection of proteins, and include isomerases, epimerases, oxidoreductases, and lyases; they typically have a TGXXGXXG cofactor binding motif. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold, an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Sequence identity between different SDR enzymes is typically in the 15-30% range; they catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase numbering). In addition to the Tyr and Lys, there is often an upstream Ser and/or an Asn, contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Atypical SDRs generally lack the catalytic residues characteristic of the SDRs, and their glycine-rich NAD(P)-binding motif is often different from the forms normally seen in classical or extended SDRs. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 15 | 1143 | 17 | 1129 | |
| 0.0 | 15 | 1141 | 17 | 1127 | |
| 0.0 | 3 | 1143 | 6 | 1131 | |
| 0.0 | 3 | 1143 | 6 | 1124 | |
| 0.0 | 3 | 1143 | 6 | 1124 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.23e-11 | 90 | 427 | 2 | 322 | Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis],2C20_B Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis],2C20_C Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis],2C20_D Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis],2C20_E Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis],2C20_F Crystal Structure Of Udp-Glucose 4-Epimerase [Bacillus anthracis] |
|
| 2.30e-09 | 88 | 412 | 2 | 305 | Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) from Hyperthermophilic Eubacterium Thermotoga maritima [Thermotoga maritima MSB8],4ZRM_B Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) from Hyperthermophilic Eubacterium Thermotoga maritima [Thermotoga maritima MSB8],4ZRN_A Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) with UDP-glucose from Hyperthermophilic Eubacterium Thermotoga Maritima [Thermotoga maritima MSB8],4ZRN_B Crystal Structure of UDP-Glucose 4-Epimerase (TM0509) with UDP-glucose from Hyperthermophilic Eubacterium Thermotoga Maritima [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.69e-20 | 821 | 1115 | 80 | 381 | Probable glucuronosyltransferase Os01g0926400 OS=Oryza sativa subsp. japonica OX=39947 GN=Os01g0926400 PE=2 SV=1 |
|
| 2.65e-18 | 820 | 1115 | 116 | 424 | Probable glucuronoxylan glucuronosyltransferase IRX7 OS=Arabidopsis thaliana OX=3702 GN=IRX7 PE=2 SV=1 |
|
| 4.76e-17 | 822 | 1115 | 74 | 374 | Probable glucuronosyltransferase Os01g0926600 OS=Oryza sativa subsp. japonica OX=39947 GN=Os01g0926600 PE=2 SV=1 |
|
| 2.08e-15 | 822 | 1115 | 79 | 379 | Probable glucuronosyltransferase Os04g0398600 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0398600 PE=2 SV=2 |
|
| 2.30e-15 | 784 | 1115 | 54 | 392 | Probable glucuronosyltransferase Os02g0520750 OS=Oryza sativa subsp. japonica OX=39947 GN=Os02g0520750 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
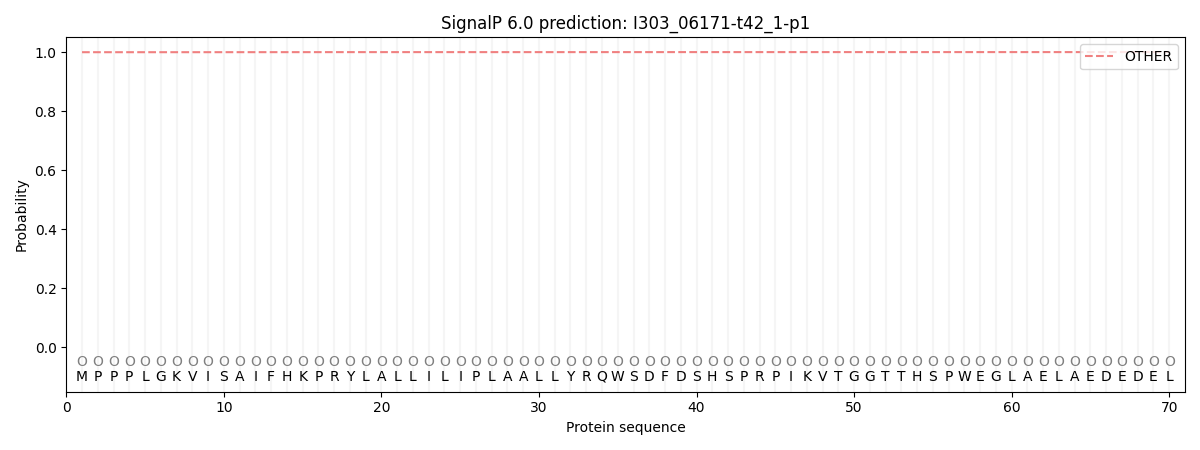
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999702 | 0.000325 |
