You are browsing environment: FUNGIDB
CAZyme Information: I303_06089-t42_1-p1
You are here: Home > Sequence: I303_06089-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
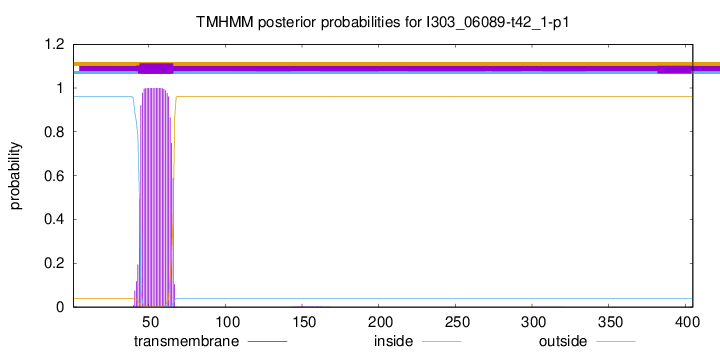
TMHMM annotations
Basic Information help
| Species | Kwoniella dejecticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella dejecticola | |||||||||||
| CAZyme ID | I303_06089-t42_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT17 | 105 | 398 | 1e-78 | 0.9823943661971831 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398414 | Glyco_transf_17 | 3.06e-127 | 56 | 404 | 15 | 349 | Glycosyltransferase family 17. This family represents beta-1,4-mannosyl-glycoprotein beta-1,4-N-acetylglucosaminyltransferase (EC:2.4.1.144). This enzyme transfers the bisecting GlcNAc to the core mannose of complex N-glycans. The addition of this residue is regulated during development and has functional consequences for receptor signalling, cell adhesion, and tumor progression. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.30e-98 | 140 | 405 | 1 | 264 | |
| 2.21e-84 | 70 | 403 | 48 | 367 | |
| 3.67e-84 | 68 | 403 | 38 | 359 | |
| 3.27e-82 | 17 | 403 | 4 | 381 | |
| 4.61e-82 | 34 | 403 | 3 | 359 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.21e-09 | 118 | 404 | 202 | 505 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Homo sapiens OX=9606 GN=MGAT3 PE=1 SV=3 |
|
| 3.92e-09 | 118 | 405 | 206 | 510 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Rattus norvegicus OX=10116 GN=Mgat3 PE=1 SV=2 |
|
| 9.17e-09 | 118 | 405 | 206 | 510 | Beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase OS=Mus musculus OX=10090 GN=Mgat3 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999980 | 0.000048 |