You are browsing environment: FUNGIDB
CAZyme Information: I303_05686-t42_1-p1
You are here: Home > Sequence: I303_05686-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
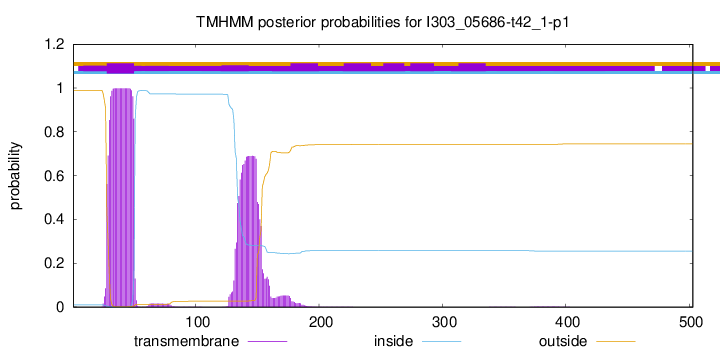
TMHMM annotations
Basic Information help
| Species | Kwoniella dejecticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella dejecticola | |||||||||||
| CAZyme ID | I303_05686-t42_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | beta-1,4-mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.142:10 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT33 | 61 | 493 | 2.6e-154 | 0.9882352941176471 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340843 | GT33_ALG1-like | 0.0 | 58 | 493 | 1 | 407 | chitobiosyldiphosphodolichol beta-mannosyltransferase and similar proteins. This family is most closely related to the GT33 family of glycosyltransferases. The yeast gene ALG1 has been shown to function as a mannosyltransferase that catalyzes the formation of dolichol pyrophosphate (Dol-PP)-GlcNAc2Man from GDP-Man and Dol-PP-Glc-NAc2, and participates in the formation of the lipid-linked precursor oligosaccharide for N-glycosylation. In humans ALG1 has been associated with the congenital disorders of glycosylation (CDG) designated as subtype CDG-Ik. |
| 215155 | PLN02275 | 1.80e-135 | 58 | 453 | 2 | 370 | transferase, transferring glycosyl groups |
| 223515 | RfaB | 4.79e-10 | 104 | 493 | 27 | 371 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
| 340816 | Glycosyltransferase_GTB-type | 4.15e-07 | 342 | 440 | 139 | 235 | glycosyltransferase family 1 and related proteins with GTB topology. Glycosyltransferases catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. The acceptor molecule can be a lipid, a protein, a heterocyclic compound, or another carbohydrate residue. The structures of the formed glycoconjugates are extremely diverse, reflecting a wide range of biological functions. The members of this family share a common GTB topology, one of the two protein topologies observed for nucleotide-sugar-dependent glycosyltransferases. GTB proteins have distinct N- and C- terminal domains each containing a typical Rossmann fold. The two domains have high structural homology despite minimal sequence homology. The large cleft that separates the two domains includes the catalytic center and permits a high degree of flexibility. |
| 340825 | GT4_WbuB-like | 8.56e-06 | 70 | 457 | 12 | 359 | Escherichia coli WbuB and similar proteins. This family is most closely related to the GT1 family of glycosyltransferases. WbuB in E. coli is involved in the biosynthesis of the O26 O-antigen. It has been proposed to function as an N-acetyl-L-fucosamine (L-FucNAc) transferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.56e-216 | 16 | 499 | 5 | 502 | |
| 3.72e-215 | 16 | 499 | 5 | 502 | |
| 3.72e-215 | 16 | 499 | 5 | 502 | |
| 7.48e-215 | 16 | 503 | 5 | 506 | |
| 2.13e-214 | 16 | 503 | 5 | 506 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-109 | 64 | 498 | 36 | 462 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Pongo abelii OX=9601 GN=ALG1 PE=2 SV=1 |
|
| 2.72e-108 | 64 | 498 | 36 | 462 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG1 PE=1 SV=2 |
|
| 2.64e-107 | 64 | 493 | 36 | 457 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Mus musculus OX=10090 GN=Alg1 PE=1 SV=3 |
|
| 7.33e-96 | 58 | 487 | 2 | 449 | UDP-glycosyltransferase TURAN OS=Arabidopsis thaliana OX=3702 GN=TUN PE=2 SV=1 |
|
| 3.56e-83 | 61 | 458 | 2 | 418 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Dictyostelium discoideum OX=44689 GN=alg1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000061 | 0.000003 |