You are browsing environment: FUNGIDB
CAZyme Information: I303_04645-t42_1-p1
You are here: Home > Sequence: I303_04645-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella dejecticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella dejecticola | |||||||||||
| CAZyme ID | I303_04645-t42_1-p1 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371727 | Glyco_hydro_cc | 1.94e-51 | 185 | 405 | 14 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.61e-78 | 168 | 413 | 7 | 254 | |
| 1.47e-77 | 168 | 413 | 7 | 254 | |
| 3.48e-77 | 168 | 413 | 23 | 270 | |
| 4.40e-75 | 168 | 413 | 23 | 270 | |
| 4.18e-74 | 168 | 413 | 7 | 254 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.81e-102 | 167 | 413 | 197 | 444 | Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
|
| 1.95e-79 | 168 | 413 | 29 | 276 | Chain A, Glyco_hydro_cc domain-containing protein [Trichoderma gamsii] |
|
| 3.70e-17 | 166 | 408 | 88 | 345 | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) [Sorangium cellulosum So ce56],6UAX_B Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Sorangium cellulosum (ScGH128_II) [Sorangium cellulosum So ce56] |
|
| 2.93e-15 | 171 | 380 | 23 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei],6UAR_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose [Amycolatopsis mediterranei] |
|
| 7.30e-15 | 171 | 380 | 23 | 234 | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose [Amycolatopsis mediterranei],6UFZ_A Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) [Amycolatopsis mediterranei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.04e-14 | 175 | 407 | 304 | 530 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
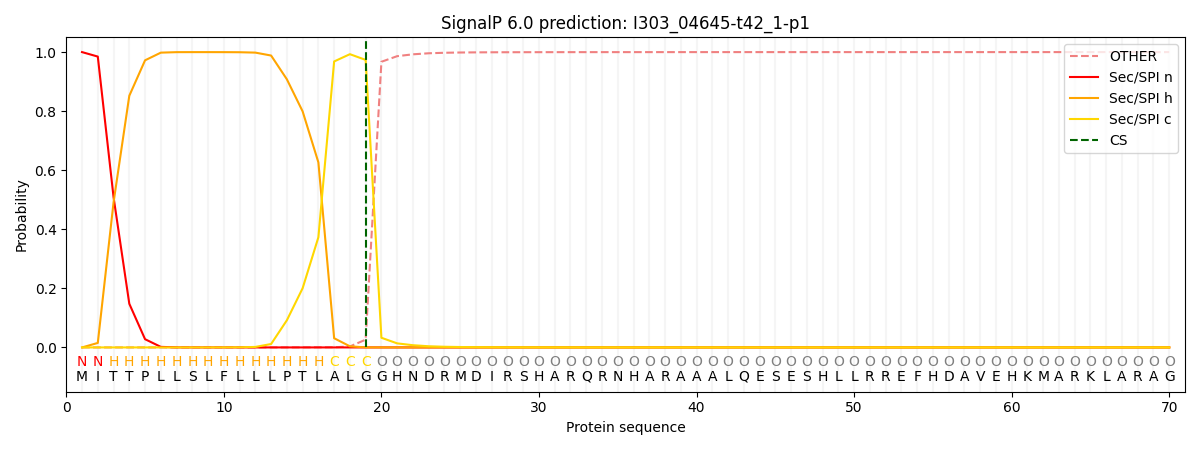
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000381 | 0.999616 | CS pos: 19-20. Pr: 0.9736 |
