You are browsing environment: FUNGIDB
CAZyme Information: I303_00203-t42_1-p1
You are here: Home > Sequence: I303_00203-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
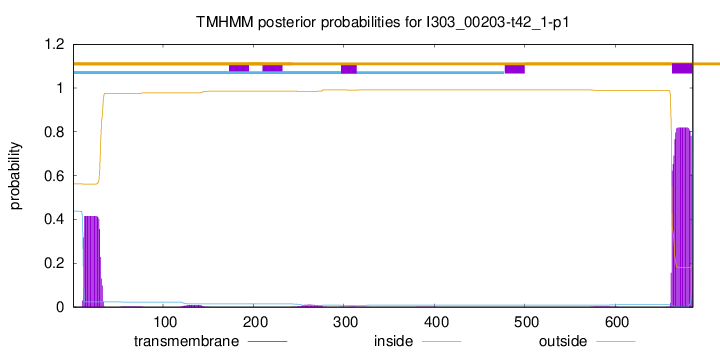
TMHMM annotations
Basic Information help
| Species | Kwoniella dejecticola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella dejecticola | |||||||||||
| CAZyme ID | I303_00203-t42_1-p1 | |||||||||||
| CAZy Family | AA11 | |||||||||||
| CAZyme Description | glyoxal oxidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA5 | 56 | 620 | 7e-228 | 0.9875666074600356 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 399910 | Glyoxal_oxid_N | 4.68e-36 | 110 | 327 | 51 | 243 | Glyoxal oxidase N-terminus. This family represents the N-terminus (approximately 300 residues) of a number of plant and fungal glyoxal oxidase enzymes. Glyoxal oxidase catalyzes the oxidation of aldehydes to carboxylic acids, coupled with reduction of dioxygen to hydrogen peroxide. It is an essential component of the extracellular lignin degradation pathways of the wood-rot fungus Phanerochaete chrysosporium. |
| 401164 | DUF1929 | 1.13e-24 | 501 | 600 | 3 | 91 | Domain of unknown function (DUF1929). Members of this family adopt a secondary structure consisting of a bundle of seven, mostly antiparallel, beta-strands surrounding a hydrophobic core. The 7 strands are arranged in 2 sheets, in a Greek-key topology. Their precise function, has not, as yet, been defined, though they are mostly found in sugar-utilising enzymes, such as galactose oxidase. |
| 199882 | E_set_GO_C | 2.31e-22 | 488 | 600 | 1 | 103 | C-terminal Early set domain associated with the catalytic domain of galactose oxidase. E or "early" set domains are associated with the catalytic domain of galactose oxidase at the C-terminal end. Galactose oxidase is an extracellular monomeric enzyme which catalyzes the stereospecific oxidation of a broad range of primary alcohol substrates and possesses a unique mononuclear copper site essential for catalyzing a two-electron transfer reaction during the oxidation of primary alcohols to corresponding aldehydes. The second redox active center necessary for the reaction was found to be situated at a tyrosine residue. The C-terminal domain of galactose oxidase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 40 | 633 | 22 | 616 | |
| 0.0 | 40 | 633 | 22 | 616 | |
| 0.0 | 40 | 633 | 22 | 616 | |
| 0.0 | 40 | 633 | 22 | 616 | |
| 1.46e-238 | 58 | 623 | 41 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.83e-13 | 195 | 596 | 253 | 608 | Structure of Galactose Oxidase homologue from Streptomyces lividans [Streptomyces lividans],4UNM_B Structure of Galactose Oxidase homologue from Streptomyces lividans [Streptomyces lividans] |
|
| 8.76e-12 | 195 | 596 | 242 | 597 | W288A mutant of GlxA from Streptomyces lividans: Cu-bound form [Streptomyces lividans TK24] |
|
| 8.82e-12 | 195 | 596 | 247 | 602 | W288A mutant of GlxA from Streptomyces lividans: apo form [Streptomyces lividans 1326],5LQI_B W288A mutant of GlxA from Streptomyces lividans: apo form [Streptomyces lividans 1326] |
|
| 6.75e-06 | 375 | 600 | 439 | 637 | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
|
| 6.75e-06 | 375 | 600 | 439 | 637 | Glactose oxidase C383S mutant identified by directed evolution [Fusarium sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.26e-69 | 67 | 603 | 35 | 552 | Aldehyde oxidase GLOX OS=Phanerodontia chrysosporium OX=2822231 GN=GLX PE=1 SV=1 |
|
| 6.93e-46 | 111 | 600 | 80 | 521 | Aldehyde oxidase GLOX OS=Vitis pseudoreticulata OX=231512 GN=GLOX PE=2 SV=1 |
|
| 1.33e-37 | 111 | 600 | 162 | 614 | Aldehyde oxidase GLOX1 OS=Arabidopsis thaliana OX=3702 GN=GLOX1 PE=2 SV=1 |
|
| 9.61e-36 | 146 | 600 | 140 | 593 | Putative aldehyde oxidase Art an 7 OS=Artemisia annua OX=35608 PE=1 SV=1 |
|
| 2.38e-34 | 120 | 617 | 477 | 922 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.015499 | 0.984485 | CS pos: 27-28. Pr: 0.5671 |