You are browsing environment: FUNGIDB
CAZyme Information: I302_08444-t42_1-p1
You are here: Home > Sequence: I302_08444-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Kwoniella bestiolae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Tremellomycetes; ; Cryptococcaceae; Kwoniella; Kwoniella bestiolae | |||||||||||
| CAZyme ID | I302_08444-t42_1-p1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 51 | 364 | 1.1e-90 | 0.9869706840390879 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350098 | GH43-like | 1.24e-04 | 148 | 259 | 46 | 150 | Glycosyl hydrolase family 43. This glycosyl hydrolase family 43 (GH43)-like subfamily includes uncharacterized enzymes similar to those with beta-1,4-xylosidase (xylan 1,4-beta-xylosidase; EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanase and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 271234 | Sialidase_non-viral | 0.007 | 155 | 372 | 52 | 275 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| 271234 | Sialidase_non-viral | 0.009 | 88 | 211 | 227 | 330 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.92e-115 | 11 | 388 | 10 | 383 | |
| 1.10e-107 | 6 | 384 | 21 | 395 | |
| 3.84e-105 | 8 | 384 | 24 | 396 | |
| 5.27e-105 | 34 | 384 | 52 | 395 | |
| 2.85e-104 | 9 | 384 | 24 | 394 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.00e-90 | 22 | 367 | 3 | 341 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
|
| 1.13e-89 | 22 | 367 | 3 | 341 | Chain A, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
|
| 1.60e-89 | 22 | 367 | 3 | 341 | Chain A, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
|
| 5.09e-88 | 22 | 367 | 3 | 341 | Chain A, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
|
| 5.65e-84 | 44 | 355 | 15 | 320 | High resolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
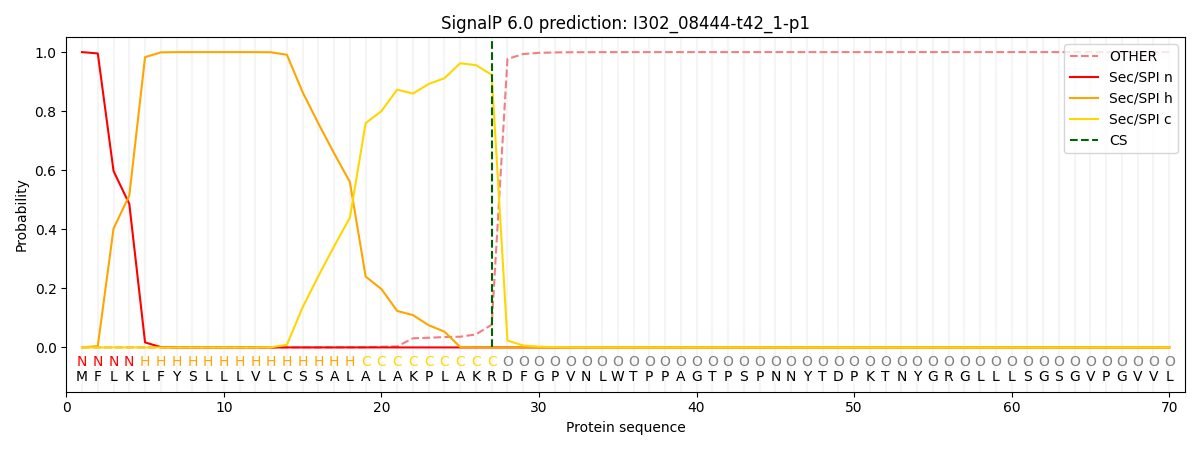
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000689 | 0.999294 | CS pos: 27-28. Pr: 0.9230 |
